4KFO
 
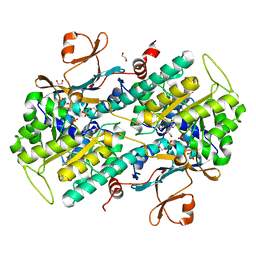 | | Structure-Based Discovery of Novel Amide-Containing Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | 分子名称: | 1,2-ETHANEDIOL, N-{4-[(3,5-difluorophenyl)sulfonyl]benzyl}imidazo[1,2-a]pyridine-6-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | 著者 | Zheng, X, Bauer, P, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynolds, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Gunzner-Tosteb, J, Liederer, B.M, Ly, J, O'Brien, T, Oh, A, Wang, L, Wang, W, Xiao, Y, Zak, M, Zhao, G, Yuen, P, Bair, K.W. | | 登録日 | 2013-04-27 | | 公開日 | 2013-05-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure-based identification of ureas as novel nicotinamide phosphoribosyltransferase (nampt) inhibitors.
J.Med.Chem., 56, 2013
|
|
4KFN
 
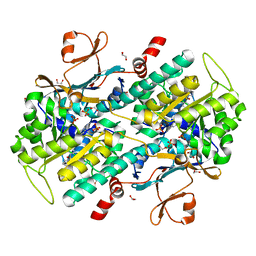 | | Structure-Based Discovery of Novel Amide-Containing Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | 分子名称: | 1,2-ETHANEDIOL, N-[4-(piperidin-1-ylsulfonyl)benzyl]-1H-pyrrolo[3,2-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | 著者 | Zheng, X, Bauer, P, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynolds, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Gunzner-Toste, J, Liederer, B.M, Ly, J, O'Brien, T, Oh, A, Wang, L, Wang, W, Xiao, Y, Zak, M, Zhao, G, Yuen, P, Bair, K.W. | | 登録日 | 2013-04-27 | | 公開日 | 2013-05-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure-based identification of ureas as novel nicotinamide phosphoribosyltransferase (nampt) inhibitors.
J.Med.Chem., 56, 2013
|
|
6JBJ
 
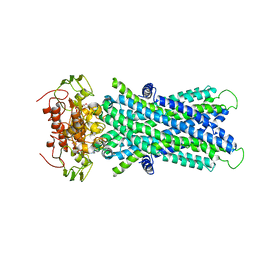 | | Cryo-EM structure of human lysosomal cobalamin exporter ABCD4 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family D member 4 | | 著者 | Xu, D, Feng, Z, Hou, W.T, Jiang, Y.L, Wang, L, Sun, L.F, Zhou, C.Z, Chen, Y. | | 登録日 | 2019-01-25 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structure of human lysosomal cobalamin exporter ABCD4.
Cell Res., 29, 2019
|
|
7DRI
 
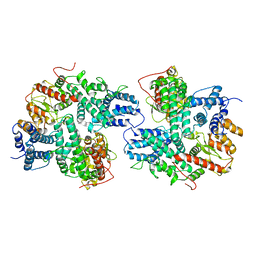 | | Structure of SspE_CTD_41658 | | 分子名称: | DUF1524 domain | | 著者 | Haiyan, G, Jinchuan, Z, Chen, S, Wang, L, Wu, G. | | 登録日 | 2020-12-28 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | Nicking mechanism underlying the DNA phosphorothioate-sensing antiphage defense by SspE.
Nat Commun, 13, 2022
|
|
4M02
 
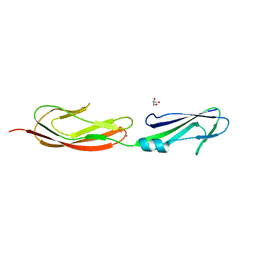 | | Middle fragment(residues 494-663) of the binding region of SraP | | 分子名称: | CALCIUM ION, GLYCEROL, Serine-rich adhesin for platelets | | 著者 | Yang, Y.H, Jiang, Y.L, Zhang, J, Wang, L, Chen, Y, Zhou, C.Z. | | 登録日 | 2013-08-01 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Structural Insights into SraP-Mediated Staphylococcus aureus Adhesion to Host Cells
Plos Pathog., 10, 2014
|
|
5W0Z
 
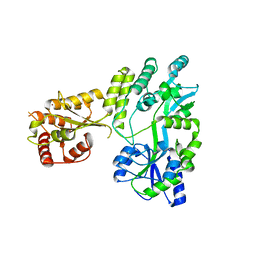 | |
4M03
 
 | | C-terminal fragment(residues 576-751) of binding region of SraP | | 分子名称: | CALCIUM ION, Serine-rich adhesin for platelets | | 著者 | Yang, Y.H, Jiang, Y.L, Zhang, J, Wang, L, Chen, Y, Zhou, C.Z. | | 登録日 | 2013-08-01 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Structural Insights into SraP-Mediated Staphylococcus aureus Adhesion to Host Cells
Plos Pathog., 10, 2014
|
|
5W0R
 
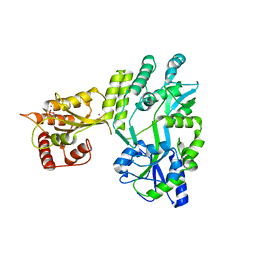 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with cacodylic acid | | 分子名称: | CACODYLATE ION, CALCIUM ION, MBP fused activation-induced cytidine deaminase, ... | | 著者 | Qiao, Q, Wang, L, Wu, H. | | 登録日 | 2017-05-31 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
4M01
 
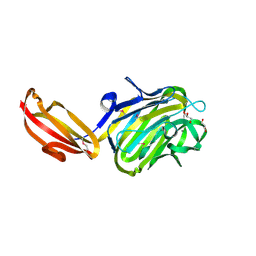 | | N terminal fragment(residues 245-575) of binding region of SraP | | 分子名称: | CALCIUM ION, GLYCEROL, Serine-rich adhesin for platelets | | 著者 | Yang, Y.H, Jiang, Y.L, Zhang, J, Wang, L, Chen, Y, Zhou, C.Z. | | 登録日 | 2013-08-01 | | 公開日 | 2014-06-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Insights into SraP-Mediated Staphylococcus aureus Adhesion to Host Cells
Plos Pathog., 10, 2014
|
|
5W0U
 
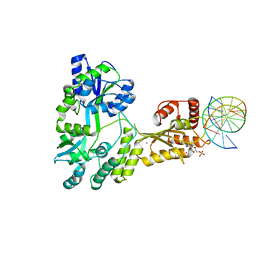 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with dCMP | | 分子名称: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*TP*GP*AP*AP*C)-3'), ... | | 著者 | Qiao, Q, Wang, L, Wu, H. | | 登録日 | 2017-05-31 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
5W1C
 
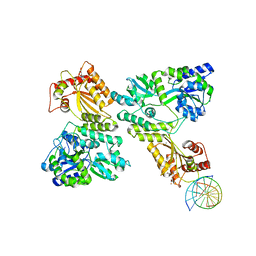 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with cytidine | | 分子名称: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*TP*GP*AP*AP*C)-3'), ... | | 著者 | Qiao, Q, Wang, L, Wu, H. | | 登録日 | 2017-06-02 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.18 Å) | | 主引用文献 | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
4M00
 
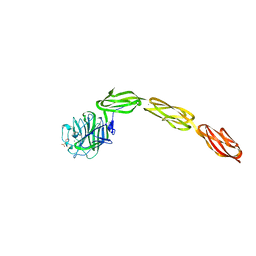 | | Crystal structure of the ligand binding region of staphylococcal adhesion SraP | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Serine-rich adhesin for platelets, ... | | 著者 | Yang, Y.H, Jiang, Y.L, Zhang, J, Wang, L, Chen, Y, Zhou, C.Z. | | 登録日 | 2013-08-01 | | 公開日 | 2014-06-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural Insights into SraP-Mediated Staphylococcus aureus Adhesion to Host Cells
Plos Pathog., 10, 2014
|
|
4N0G
 
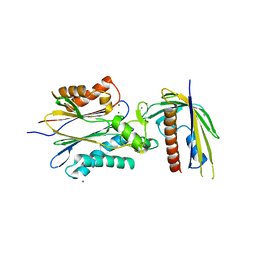 | | Crystal Structure of PYL13-PP2CA complex | | 分子名称: | Abscisic acid receptor PYL13, MAGNESIUM ION, Protein phosphatase 2C 37, ... | | 著者 | Li, W, Wang, L, Sheng, X, Yan, C, Zhou, R, Hang, J, Yin, P, Yan, N. | | 登録日 | 2013-10-01 | | 公開日 | 2013-11-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.382 Å) | | 主引用文献 | Molecular basis for the selective and ABA-independent inhibition of PP2CA by PYL13
Cell Res., 23, 2013
|
|
4MZ4
 
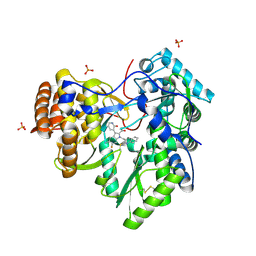 | | Discovery of an Irreversible HCV NS5B Polymerase Inhibitor | | 分子名称: | 1-[(2-chloroquinolin-3-yl)methyl]-6-fluoro-5-methyl-3-(2-oxo-1,2-dihydropyridin-3-yl)-1H-indole-2-carboxylic acid, PHOSPHATE ION, RNA-directed RNA polymerase | | 著者 | Zeng, Q, Anilkumar, G.N, Rosenblum, S.B, Huang, H.-C, Lesburg, C.A, Jiang, Y, Selyutin, O, Chan, T.-Y, Bennett, F, Chen, K.X, Venkatraman, S, Sannigrahi, M, Velazquez, F, Duca, J.S, Gavalas, S, Huang, Y, Pu, H, Wang, L, Pinto, P, Vibulbhan, B, Agrawal, S, Ferrari, E, Jiang, C.-K, Li, C, Hesk, D, Gesell, J, Sorota, S, Shih, N.-Y, Njoroge, F.G, Kozlowski, J.A. | | 登録日 | 2013-09-29 | | 公開日 | 2013-12-11 | | 最終更新日 | 2013-12-18 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Discovery of an irreversible HCV NS5B polymerase inhibitor.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5XM5
 
 | |
4OJR
 
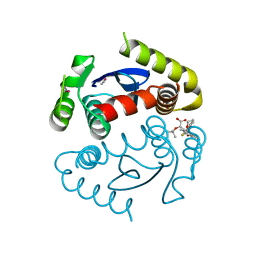 | | Crystal Structure of the HIV-1 Integrase catalytic domain with GSK1264 | | 分子名称: | (2S)-tert-butoxy[4-(8-fluoro-5-methyl-3,4-dihydro-2H-chromen-6-yl)-2-methyl-1-oxo-1,2-dihydroisoquinolin-3-yl]ethanoic acid, CACODYLATE ION, HIV-1 Integrase, ... | | 著者 | Gupta, K, Brady, T, Dyer, B, Hwang, Y, Male, F, Nolte, R.T, Wang, L, Velthuisen, E, Jeffrey, J, Van Duyne, G, Bushman, F.D. | | 登録日 | 2014-01-21 | | 公開日 | 2014-06-11 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Allosteric Inhibition of Human Immunodeficiency Virus Integrase: LATE BLOCK DURING VIRAL REPLICATION AND ABNORMAL MULTIMERIZATION INVOLVING SPECIFIC PROTEIN DOMAINS.
J.Biol.Chem., 289, 2014
|
|
5YXC
 
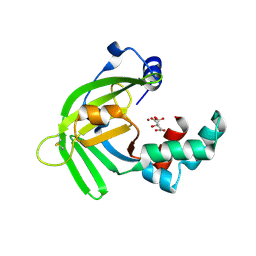 | | Crystal structure of Zinc binding protein ZinT in complex with citrate from E. coli | | 分子名称: | CITRIC ACID, Metal-binding protein ZinT, ZINC ION | | 著者 | Chen, J, Wang, L, Shang, F, Xu, Y. | | 登録日 | 2017-12-04 | | 公開日 | 2017-12-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.763 Å) | | 主引用文献 | Crystal structure of E. coli ZinT with one zinc-binding mode and complexed with citrate
Biochem. Biophys. Res. Commun., 500, 2018
|
|
5Z7H
 
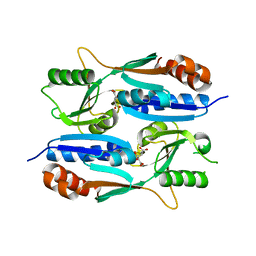 | |
4PIV
 
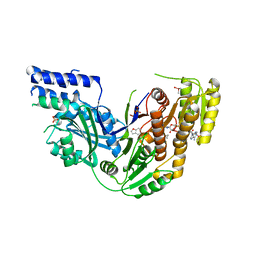 | | Human Fatty Acid Synthase Psi/KR Tri-Domain with NADPH and GSK2194069 | | 分子名称: | 4-[4-(1-benzofuran-5-yl)phenyl]-5-{[(3S)-1-(cyclopropylcarbonyl)pyrrolidin-3-yl]methyl}-2,4-dihydro-3H-1,2,4-triazol-3-one, CACODYLATE ION, Fatty acid synthase, ... | | 著者 | Williams, S.P, Wang, L, Brown, K.K, Parrish, C.A, Hardwicke, M.A. | | 登録日 | 2014-05-09 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.299 Å) | | 主引用文献 | A human fatty acid synthase inhibitor binds beta-ketoacyl reductase in the keto-substrate site.
Nat.Chem.Biol., 10, 2014
|
|
5Z72
 
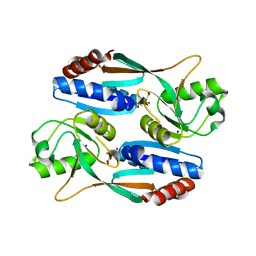 | |
7YSK
 
 | | Crystal structure of D-Cysteine desulfhydrase from Pectobacterium atrosepticum | | 分子名称: | D-Cysteine desulfhydrase | | 著者 | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | 登録日 | 2022-08-12 | | 公開日 | 2022-12-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
7YSL
 
 | | Crystal structure of D-Cysteine desulfhydrase with a trapped PLP-pyruvate geminal diamine | | 分子名称: | 1,2-ETHANEDIOL, D-Cysteine desulfhydrase, FORMIC ACID | | 著者 | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | 登録日 | 2022-08-12 | | 公開日 | 2022-12-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
7DRS
 
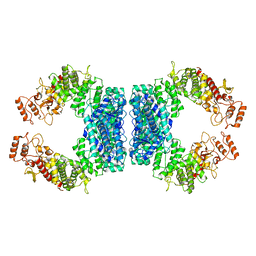 | | Structure of SspE_40224 | | 分子名称: | SspE protein | | 著者 | Haiyan, G, Jinchuan, Z, Chen, S, Wang, L, Wu, G. | | 登録日 | 2020-12-29 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.42 Å) | | 主引用文献 | Nicking mechanism underlying the DNA phosphorothioate-sensing antiphage defense by SspE.
Nat Commun, 13, 2022
|
|
7DRR
 
 | | Structure of SspE-R100A protein | | 分子名称: | SspE protein | | 著者 | Haiyan, G, Jinchuan, Z, Chen, S, Wang, L, Wu, G. | | 登録日 | 2020-12-29 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.48 Å) | | 主引用文献 | Nicking mechanism underlying the DNA phosphorothioate-sensing antiphage defense by SspE.
Nat Commun, 13, 2022
|
|
7E5S
 
 | | SARS-CoV-2 S trimer with four-antibody cocktail complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FC05 heavy chain, FC05 light chain, ... | | 著者 | Sun, Y, Wang, L, Wang, N, Feng, R, Wang, X. | | 登録日 | 2021-02-20 | | 公開日 | 2021-11-17 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure-based development of three- and four-antibody cocktails against SARS-CoV-2 via multiple mechanisms.
Cell Res., 31, 2021
|
|
