4GAZ
 
 | | Crystal Structure of a Jumonji Domain-containing Protein JMJD5 | | Descriptor: | Lysine-specific demethylase 8, N-OXALYLGLYCINE, NICKEL (II) ION | | Authors: | Wang, H, Zhou, X, Zhang, X, Tao, Y, Chen, N, Zang, J. | | Deposit date: | 2012-07-26 | | Release date: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal Structure of a Jumonji Domain-containing Protein JMJD5
To be Published
|
|
4FY0
 
 | |
2YVR
 
 | | Crystal structure of MS1043 | | Descriptor: | Transcription intermediary factor 1-beta, ZINC ION | | Authors: | Wang, H, Kishishita, S, Murayama, K, Takemoto, C, Terada, T, Shirouzu, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-13 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of MS1043
To be Published
|
|
2Z0I
 
 | | Crystal Structure of 5-aminolevulinic acid dehydratase (ALAD) from Mus musculus | | Descriptor: | Delta-aminolevulinic acid dehydratase | | Authors: | Wang, H, Xie, Y, Kawazoe, M, Kishishita, S, Murayama, K, Takemoto, C, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2008-05-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of 5-aminolevulinic acid dehydratase (ALAD) from Mus musculus
To be Published
|
|
4HN2
 
 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with AMPPNP and a substrate analog 5PA-IP5 | | Descriptor: | (2-oxo-2-{[(1s,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl]oxy}ethyl)phosphonic acid, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase 2, MAGNESIUM ION, ... | | Authors: | Wang, H. | | Deposit date: | 2012-10-18 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | First synthetic analogues of diphosphoinositol polyphosphates: interaction with PP-InsP(5) kinase.
Chem.Commun.(Camb.), 48, 2012
|
|
8HEI
 
 | | Crystal structure of CTSB in complex with E64d | | Descriptor: | Cathepsin B, GLYCEROL, ethyl (3S)-3-hydroxy-4-({(2S)-4-methyl-1-[(3-methylbutyl)amino]-1-oxopentan-2-yl}amino)-4-oxobutanoate | | Authors: | Wang, H, Li, D, Sun, L, Yang, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HET
 
 | | Crystal structure of CTSL in complex with E64d | | Descriptor: | Procathepsin L, ethyl (3S)-3-hydroxy-4-({(2S)-4-methyl-1-[(3-methylbutyl)amino]-1-oxopentan-2-yl}amino)-4-oxobutanoate | | Authors: | Wang, H, Shao, M, Sun, L, Yang, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HD8
 
 | | Crystal structure of TMPRSS2 in complex with 212-148 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, CALCIUM ION, ... | | Authors: | Wang, H, Liu, X, Sun, L, Yang, H. | | Deposit date: | 2022-11-03 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HE9
 
 | | Crystal structure of CTSB in complex with K777 | | Descriptor: | Cathepsin B, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Wang, H, Li, D, Sun, L, Yang, H. | | Deposit date: | 2022-11-07 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HFV
 
 | | Crystal structure of CTSL in complex with K777 | | Descriptor: | CACODYLATE ION, Nalpha-[(4-methylpiperazin-1-yl)carbonyl]-N-[(3S)-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]-L-phenylalaninamide, Procathepsin L, ... | | Authors: | Wang, H, Shao, M, Sun, L, Yang, H. | | Deposit date: | 2022-11-12 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
8HEN
 
 | | Crystal structure of CTSB in complex with 212-148 | | Descriptor: | 2-[4-[[(2~{S})-1-oxidanylidene-3-phenyl-1-[[(3~{S})-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]amino]propan-2-yl]carbamoyl]piperazin-1-yl]ethyl 4-carbamimidamidobenzoate, Cathepsin B, DIMETHYL SULFOXIDE, ... | | Authors: | Wang, H, Li, D, Sun, L, Yang, H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based discovery of dual pathway inhibitors for SARS-CoV-2 entry.
Nat Commun, 14, 2023
|
|
2ZUL
 
 | | Crystal structure of Thermus thermophilus 16S rRNA methyltransferase RsmC (TTHA0533) in complex with cofactor S-adenosyl-L-Methionine | | Descriptor: | Probable ribosomal RNA small subunit methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Wang, H, Kawazoe, M, Tatsuguchi, A, Naoe, C, Takemoto, C, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-10-21 | | Release date: | 2009-10-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Thermus thermophilus 16S rRNA methyltransferase RsmC (TTHA0533) in complex with cofactor S-adenosyl-L-Methionine
To be Published
|
|
2ZWV
 
 | | Crystal structure of Thermus thermophilus 16S rRNA methyltransferase RsmC (TTHA0533) | | Descriptor: | Probable ribosomal RNA small subunit methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wang, H, Kawazoe, M, Kaminishi, T, Tatsuguchi, A, Naoe, C, Terada, T, Shirouzu, M, Takemoto, C, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-12-18 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Thermus thermophilus 16S rRNA methyltransferase RsmC (TTHA0533)
to be published
|
|
2H40
 
 | | Crystal structure of the catalytic domain of unliganded PDE5 | | Descriptor: | MAGNESIUM ION, ZINC ION, cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Wang, H, Liu, Y, Huai, Q, Cai, J, Zoraghi, R, Francis, S.H, Corbin, J.D, Robinson, H, Xin, Z, Lin, G, Ke, H. | | Deposit date: | 2006-05-23 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Multiple Conformations of Phosphodiesterase-5: Implications for enzyme function and drug development
J.Biol.Chem., 281, 2006
|
|
2H42
 
 | | Crystal structure of PDE5 in complex with sildenafil | | Descriptor: | 5-{2-ETHOXY-5-[(4-METHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-1-METHYL-3-PROPYL-1H,6H,7H-PYRAZOLO[4,3-D]PYRIMIDIN-7-ONE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wang, H, Ke, H. | | Deposit date: | 2006-05-23 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple Conformations of Phosphodiesterase-5: Implications for enzyme function and drug development
J.Biol.Chem., 281, 2006
|
|
2H44
 
 | | Crystal structure of PDE5A1 in complex with icarisid II | | Descriptor: | 5,7-DIHYDROXY-2-(4-METHOXYPHENYL)-8-(3-METHYLBUTYL)-4-OXO-4H-CHROMEN-3-YL 6-DEOXY-ALPHA-L-MANNOPYRANOSIDE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wang, H, Ke, H. | | Deposit date: | 2006-05-23 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Multiple Conformations of Phosphodiesterase-5: Implications for enzyme function and drug development
J.Biol.Chem., 281, 2006
|
|
4Q4C
 
 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with ADP and synthetic 1,5-(PP)2-IP4 (1,5-IP8) | | Descriptor: | (1R,3S,4R,5S,6R)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], ADENOSINE-5'-DIPHOSPHATE, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase 2, ... | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2014-04-14 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis of Densely Phosphorylated Bis-1,5-Diphospho-myo-Inositol Tetrakisphosphate and its Enantiomer by Bidirectional P-Anhydride Formation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5F1B
 
 | | Structural basis of Ebola virus entry: viral glycoprotein bound to its endosomal receptor Niemann-Pick C1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GP1, GP2, ... | | Authors: | Wang, H, Shi, Y, Song, J, Qi, J, Lu, G, Yan, J, Gao, G.F. | | Deposit date: | 2015-11-30 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ebola Viral Glycoprotein Bound to Its Endosomal Receptor Niemann-Pick C1.
Cell, 164, 2016
|
|
5F18
 
 | | Structural basis of Ebola virus entry: viral glycoprotein bound to its endosomal receptor Niemann-Pick C1 | | Descriptor: | Niemann-Pick C1 protein | | Authors: | Wang, H, Shi, Y, Song, J, Qi, J, Lu, G, Yan, J, Gao, G.F. | | Deposit date: | 2015-11-30 | | Release date: | 2016-01-20 | | Last modified: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ebola Viral Glycoprotein Bound to Its Endosomal Receptor Niemann-Pick C1.
Cell, 164, 2016
|
|
6HBC
 
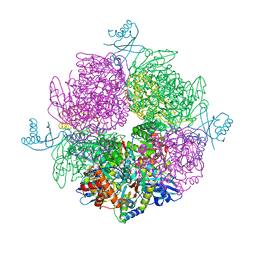 | | Structure of the repeat unit in the network formed by CcmM and Rubisco from Synechococcus elongatus | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM, Ribulose 1,5-bisphosphate carboxylase small subunit, Ribulose bisphosphate carboxylase large chain | | Authors: | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
6HBB
 
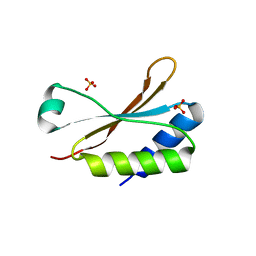 | | Crystal Structure of the small subunit-like domain 1 of CcmM from Synechococcus elongatus (strain PCC 7942) | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM, SULFATE ION | | Authors: | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
6HBA
 
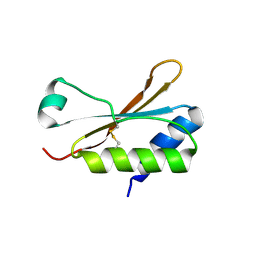 | | Crystal Structure of the small subunit-like domain 1 of CcmM from Synechococcus elongatus (strain PCC 7942), thiol-oxidized form | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM | | Authors: | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
4Q4D
 
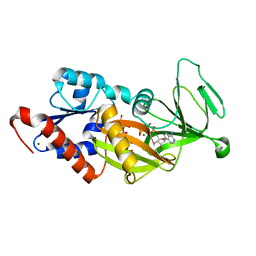 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with AMP-PNP and synthetic 3,5-(PP)2-IP4 (3,5-IP8) | | Descriptor: | (1R,3S,4S,5R,6S)-2,4,5,6-tetrakis(phosphonooxy)cyclohexane-1,3-diyl bis[trihydrogen (diphosphate)], Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase 2, MAGNESIUM ION, ... | | Authors: | Wang, H, Shears, S.B. | | Deposit date: | 2014-04-14 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis of Densely Phosphorylated Bis-1,5-Diphospho-myo-Inositol Tetrakisphosphate and its Enantiomer by Bidirectional P-Anhydride Formation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3T9E
 
 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with ADP, 5-(PP)-IP5 (5-IP7) and MgF3 (transition state mimic) | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, ADENOSINE-5'-DIPHOSPHATE, Inositol Pyrophosphate Kinase, ... | | Authors: | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | Deposit date: | 2011-08-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
3T99
 
 | | Crystal structure of the catalytic domain of human diphosphoinositol pentakisphosphate kinase 2 (PPIP5K2) in complex with ADP and in the absence of cadmium at pH 7.0 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Inositol Pyrophosphate Kinase, MAGNESIUM ION | | Authors: | Wang, H, Falck, J, Hall, T.M.T, Shears, S.B. | | Deposit date: | 2011-08-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding.
Nat.Chem.Biol., 8, 2011
|
|
