7KQT
 
 | |
4M8M
 
 | |
4FAE
 
 | | Substrate p2/NC in Complex with a Human Immunodeficiency Virus Type 1 Protease Variant | | Descriptor: | HIV-1 protease, Substrate p2/NC peptide | | Authors: | Wang, Y, Dewdney, T.G, Liu, Z, Reiter, S.J, Brunzelle, J.S, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2012-05-22 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Higher Desolvation Energy Reduces Molecular Recognition in Multi-Drug Resistant HIV-1 Protease.
Biology (Basel), 1, 2012
|
|
4K2C
 
 | | HSA Ligand Free | | Descriptor: | Serum albumin | | Authors: | Wang, Y, Luo, Z, Shi, X, Huang, M. | | Deposit date: | 2013-04-08 | | Release date: | 2013-05-01 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structural mechanism of ring-opening reaction of glucose by human serum albumin.
J. Biol. Chem., 288, 2013
|
|
2Z4B
 
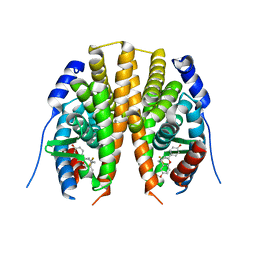 | | Estrogen receptor beta ligand-binding domain complexed to a benzopyran ligand | | Descriptor: | (3AS,4R,9BR)-2,2-DIFLUORO-4-(4-HYDROXYPHENYL)-1,2,3,3A,4,9B-HEXAHYDROCYCLOPENTA[C]CHROMEN-8-OL, Estrogen receptor beta | | Authors: | Wang, Y. | | Deposit date: | 2007-06-14 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Benzopyrans as selective estrogen receptor beta agonists (SERBAs). Part 3: Synthesis of cyclopentanone and cyclohexanone intermediates for C-ring modification.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
4FAF
 
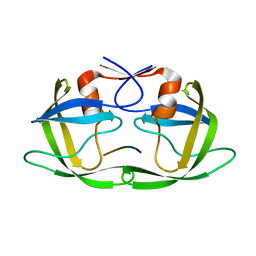 | | Substrate CA/p2 in Complex with a Human Immunodeficiency Virus Type 1 Protease Variant | | Descriptor: | HIV-1 protease, substrate CA/p2 peptide | | Authors: | Wang, Y, Dewdney, T.G, Liu, Z, Reiter, S.J, Brunzelle, J.S, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2012-05-22 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Higher Desolvation Energy Reduces Molecular Recognition in Multi-Drug Resistant HIV-1 Protease.
Biology (Basel), 1, 2012
|
|
3SO9
 
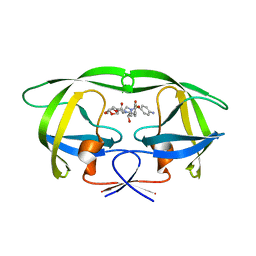 | | Darunavir in Complex with a Human Immunodeficiency Virus Type 1 Protease Variant | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HIV-1 protease | | Authors: | Wang, Y, Liu, Z, Brunzelle, S.J, Kovari, L.C, Kovari, I.A. | | Deposit date: | 2011-06-30 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | The higher barrier of darunavir and tipranavir resistance for HIV-1 protease.
Biochem.Biophys.Res.Commun., 412, 2011
|
|
3QGB
 
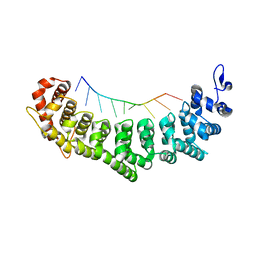 | | Crystal structure of FBF-2 R288Y mutant in complex with gld-1 FBEa | | Descriptor: | 5'-R(*UP*GP*UP*GP*CP*CP*AP*UP*A)-3', Fem-3 mRNA-binding factor 2 | | Authors: | Wang, Y, Qiu, C, Koh, Y.Y, Opperman, L, Gross, L, Hall, T.M.T, Wickens, M. | | Deposit date: | 2011-01-24 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Stacking interactions in PUF-RNA complexes.
Rna, 17, 2011
|
|
4JFK
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with (1S,6R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-10-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,6R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-10-[(2-oxo-2,3-dihydro-1,3-benzothiazol-6-yl)sulfonyl]-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4IW2
 
 | | HSA-glucose complex | | Descriptor: | D-glucose, PHOSPHATE ION, Serum albumin, ... | | Authors: | Wang, Y, Yu, H, Shi, X, Luo, Z, Huang, M. | | Deposit date: | 2013-01-23 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural mechanism of ring-opening reaction of glucose by human serum albumin
J.Biol.Chem., 288, 2013
|
|
6BLQ
 
 | | Crystal Structure of IAg7 in complex with insulin mimotope p8E9E | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2017-11-11 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C-terminal modification of the insulin B:11-23 peptide creates superagonists in mouse and human type 1 diabetes.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4P5M
 
 | | Structural Basis of Chronic Beryllium Disease: Bridging the Gap Between Allergic Hypersensitivity and Autoimmunity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)][2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Y, Dai, S, Kappler, J. | | Deposit date: | 2014-03-18 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of chronic beryllium disease: linking allergic hypersensitivity and autoimmunity.
Cell, 158, 2014
|
|
4PES
 
 | |
4PF7
 
 | | Crystal structure of insulin degrading enzyme complexed with inhibitor | | Descriptor: | (2S)-2-amino-N-{(1S)-1-cyclohexyl-2-[(4-methylphenyl)amino]-2-oxoethyl}-4-(methylselanyl)butanamide, Insulin-degrading enzyme, ZINC ION | | Authors: | Wang, Y, Guo, S. | | Deposit date: | 2014-04-28 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Dual Exosite-binding Inhibitors of Insulin-degrading Enzyme Challenge Its Role as the Primary Mediator of Insulin Clearance in Vivo.
J.Biol.Chem., 290, 2015
|
|
6CCY
 
 | | Crystal structure of Akt1 in complex with a selective inhibitor | | Descriptor: | (5R)-4-(4-{4-[4-fluoro-3-(trifluoromethyl)phenyl]-1-[2-(pyrrolidin-1-yl)ethyl]-1H-imidazol-2-yl}piperidin-1-yl)-5-methyl-5,8-dihydropyrido[2,3-d]pyrimidin-7(6H)-one, RAC-alpha serine/threonine-protein kinase,PIFtide | | Authors: | Wang, Y, Stout, S. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-02 | | Last modified: | 2018-05-16 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of chiral dihydropyridopyrimidinones as potent, selective and orally bioavailable inhibitors of AKT.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
4PFC
 
 | | Crystal structure of insulin degrading enzyme complexed with inhibitor | | Descriptor: | Insulin-degrading enzyme, ZINC ION, methyl [(2S)-2-(5-{5-[4-({(2S)-2-[(3S)-3-amino-2-oxopiperidin-1-yl]-2-cyclohexylacetyl}amino)phenyl]pentyl}-2-fluorophenyl)-3-(quinolin-3-yl)propyl]carbamate | | Authors: | Wang, Y, Guo, S. | | Deposit date: | 2014-04-28 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Dual Exosite-binding Inhibitors of Insulin-degrading Enzyme Challenge Its Role as the Primary Mediator of Insulin Clearance in Vivo.
J.Biol.Chem., 290, 2015
|
|
6BLX
 
 | | Crystal structure of IAg7 in complex with insulin mimotope p8G9E | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2017-11-11 | | Release date: | 2017-12-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.323 Å) | | Cite: | C-terminal modification of the insulin B:11-23 peptide creates superagonists in mouse and human type 1 diabetes.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5H00
 
 | |
4PF9
 
 | | Crystal structure of insulin degrading enzyme complexed with inhibitor | | Descriptor: | Insulin-degrading enzyme, ZINC ION, methyl [(2S)-2-[4-({5-[4-({(2S)-2-[(3S)-3-amino-2-oxopiperidin-1-yl]-2-cyclohexylacetyl}amino)phenyl]pentyl}oxy)phenyl]-3-(quinolin-3-yl)propyl]carbamate | | Authors: | Wang, Y, Guo, S. | | Deposit date: | 2014-04-28 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dual Exosite-binding Inhibitors of Insulin-degrading Enzyme Challenge Its Role as the Primary Mediator of Insulin Clearance in Vivo.
J.Biol.Chem., 290, 2015
|
|
6BLR
 
 | | Crystal Structure of IAg7 in complex with insulin mimotope p8E9E6SS | | Descriptor: | 1,2-ETHANEDIOL, H2-Ab1 protein, IAg7 alpha chain | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2017-11-11 | | Release date: | 2017-12-20 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | C-terminal modification of the insulin B:11-23 peptide creates superagonists in mouse and human type 1 diabetes.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5GZX
 
 | |
5G4C
 
 | | Human SIRT2 catalyse short chain fatty acyl lysine | | Descriptor: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-2, SIRT2, ... | | Authors: | Wang, Y. | | Deposit date: | 2016-05-09 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | SIRT2 Reverses 4-Oxononanoyl Lysine Modification on Histones.
J. Am. Chem. Soc., 138, 2016
|
|
5DMK
 
 | | Crystal Structure of IAg7 in complex with RLGL-WE14 | | Descriptor: | CITRATE ANION, H-2 class II histocompatibility antigen, A-D alpha chain, ... | | Authors: | Wang, Y, Jin, N, Dai, S, Kappler, J.W. | | Deposit date: | 2015-09-08 | | Release date: | 2015-10-28 | | Last modified: | 2016-04-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | N-terminal additions to the WE14 peptide of chromogranin A create strong autoantigen agonists in type 1 diabetes.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5GZY
 
 | |
6DFX
 
 | | human diabetogenic TCR T1D3 in complex with DQ8-p8E9E peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Y, Dai, S. | | Deposit date: | 2018-05-15 | | Release date: | 2019-04-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | How C-terminal additions to insulin B-chain fragments create superagonists for T cells in mouse and human type 1 diabetes.
Sci Immunol, 4, 2019
|
|
