6JD7
 
 | | Crystal structure of anti-CRISPR protein AcrIIC2 dimer | | 分子名称: | ACETATE ION, AcrIIC2, MAGNESIUM ION | | 著者 | Cheng, Z, Huang, X, Sun, W, Wang, Y.L. | | 登録日 | 2019-01-31 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Inhibition of CRISPR-Cas9 ribonucleoprotein complex assembly by anti-CRISPR AcrIIC2.
Nat Commun, 10, 2019
|
|
6JDV
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in catalytic state | | 分子名称: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, MAGNESIUM ION, ... | | 著者 | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | 登録日 | 2019-02-02 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
2AOI
 
 | | Crystal structure analysis of HIV-1 protease with a substrate analog P1-P6 | | 分子名称: | PEPTIDE INHIBITOR, POL POLYPROTEIN, SULFATE ION | | 著者 | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2005-08-12 | | 公開日 | 2006-01-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
6JDQ
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA | | 分子名称: | CRISPR-associated endonuclease Cas9, sgRNA | | 著者 | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | 登録日 | 2019-02-02 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JE3
 
 | | Crystal structure of Nme2Cas9 in complex with sgRNA and target DNA (AGGCCC PAM) with 5 nt overhang | | 分子名称: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, non-target DNA strand, ... | | 著者 | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | 登録日 | 2019-02-03 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.931 Å) | | 主引用文献 | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
3GO7
 
 | | Crystal Structure of M. tuberculosis ribokinase (Rv2436) in complex with ribose | | 分子名称: | MAGNESIUM ION, RIBOKINASE RBSK, alpha-D-ribofuranose | | 著者 | Masters, E.I, Sun, Y, Wang, Y, Parker, W.B, Li, R. | | 登録日 | 2009-03-18 | | 公開日 | 2010-03-31 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure and biochemical characterization of M.tuberculosis ribokinase (Rv2436)
To be Published
|
|
4N76
 
 | | Structure of Thermus thermophilus Argonaute bound to guide DNA and cleaved target DNA with Mn2+ | | 分子名称: | 5'-D(P*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3', Argonaute, ... | | 著者 | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | 登録日 | 2013-10-15 | | 公開日 | 2014-01-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4N41
 
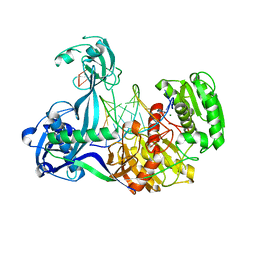 | | Structure of Thermus thermophilus Argonaute bound to guide DNA and 15-mer target DNA | | 分子名称: | 5'-D(*AP*AP*CP*CP*TP*AP*CP*TP*GP*CP*CP*TP*CP*G)-3', 5'-D(P*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*CP*G)-3', 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*T*GP*TP*AP*TP*AP*GP*T)-3', ... | | 著者 | Sheng, G, Zhao, H, Wang, J, Rao, Y, Wang, Y. | | 登録日 | 2013-10-08 | | 公開日 | 2014-01-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.248 Å) | | 主引用文献 | Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7VMA
 
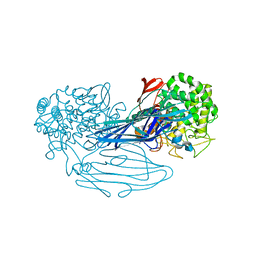 | | The X-ray crystallographic structure of amylo-alpha-1,6-glucosidase from Thermococcus gammatolerans STB12 | | 分子名称: | Amylo-alpha-1,6-glucosidase, putative archaeal type glycogen debranching enzyme (Gde) | | 著者 | Li, Z.F, Ban, X.F, Wang, Y.M, Li, C.M, Gu, Z.B. | | 登録日 | 2021-10-08 | | 公開日 | 2022-10-12 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.804 Å) | | 主引用文献 | The X-ray Crystallographic Structure of Amylo-alpha-1,6-glucosidase from Thermococcus gannatilerans STB12
To Be Published
|
|
6KC8
 
 | | Crystal structure of WT Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in post-cleavage state | | 分子名称: | CRISPR-associated endonuclease Cas9, DNA (5'-D(*AP*TP*AP*TP*GP*AP*TP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*AP*TP*CP*AP*TP*AP*TP*GP*TP*A)-3'), ... | | 著者 | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | 登録日 | 2019-06-27 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
4JKR
 
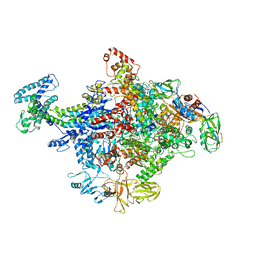 | | Crystal Structure of E. coli RNA Polymerase in complex with ppGpp | | 分子名称: | DNA-DIRECTED RNA POLYMERASE SUBUNIT BETA', DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Zuo, Y, Wang, Y, Steitz, T.A. | | 登録日 | 2013-03-11 | | 公開日 | 2013-05-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (4.2 Å) | | 主引用文献 | The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex.
Mol.Cell, 50, 2013
|
|
7CZ5
 
 | | Cryo-EM structure of the human growth hormone-releasing hormone receptor-Gs protein complex | | 分子名称: | CHOLESTEROL, Growth hormone-releasing hormone receptor,growth hormone-releasing hormone receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Zhou, F, Zhang, H, Cong, Z, Zhao, L, Zhou, Q, Mao, C, Cheng, X, Shen, D, Cai, X, Ma, C, Wang, Y, Dai, A, Zhou, Y, Sun, W, Zhao, F, Zhao, S, Jiang, H, Jiang, Y, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | 登録日 | 2020-09-07 | | 公開日 | 2020-11-18 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Structural basis for activation of the growth hormone-releasing hormone receptor.
Nat Commun, 11, 2020
|
|
5GQH
 
 | | Cryo-EM structure of PaeCas3-AcrF3 complex | | 分子名称: | CRISPR-associated nuclease/helicase Cas3 subtype I-F/YPEST, anti-CRISPR protein 3 | | 著者 | Zhang, X, Ma, J, Wang, Y, Wang, J. | | 登録日 | 2016-08-07 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | A CRISPR evolutionary arms race: structural insights into viral anti-CRISPR/Cas responses
Cell Res., 26, 2016
|
|
7ESI
 
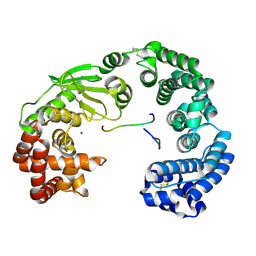 | | Crystal structure of the collagenase unit of a Vibrio collagenase from Vibrio harveyi VHJR7 at 1. 8 angstrom resolution. | | 分子名称: | CALCIUM ION, Collagenase unit (CU), Peptide P1, ... | | 著者 | Cao, H.Y, Wang, Y, Peng, M, Zhang, Y.Z. | | 登録日 | 2021-05-11 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-02-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of Vibrio collagenase VhaC provides insight into the mechanism of bacterial collagenolysis.
Nat Commun, 13, 2022
|
|
3RJX
 
 | | Crystal Structure of Hyperthermophilic Endo-Beta-1,4-glucanase | | 分子名称: | Endoglucanase FnCel5A | | 著者 | Zheng, B.S, Yang, W, Zhao, X.Y, Wang, Y.G, Lou, Z.Y, Rao, Z.H, Feng, Y. | | 登録日 | 2011-04-15 | | 公開日 | 2011-12-14 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of hyperthermophilic endo-beta-1,4-glucanase: implications for catalytic mechanism and thermostability.
J.Biol.Chem., 287, 2012
|
|
7EVP
 
 | | Cryo-EM structure of the Gp168-beta-clamp complex | | 分子名称: | Beta sliding clamp, Sliding clamp inhibitor | | 著者 | Liu, B, Li, S, Liu, Y, Chen, H, Hu, Z, Wang, Z, Gou, L, Zhang, L, Ma, B, Wang, H, Matthews, S, Wang, Y, Zhang, K. | | 登録日 | 2021-05-21 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Bacteriophage Twort protein Gp168 is a beta-clamp inhibitor by occupying the DNA sliding channel.
Nucleic Acids Res., 49, 2021
|
|
2AOH
 
 | | Crystal structure analysis of HIV-1 Protease mutant V82A with a substrate analog P6-PR | | 分子名称: | CHLORIDE ION, PEPTIDE INHIBITOR, POL POLYPROTEIN, ... | | 著者 | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Liu, F, Chen, X, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2005-08-12 | | 公開日 | 2006-01-17 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs.
Febs J., 272, 2005
|
|
1K1T
 
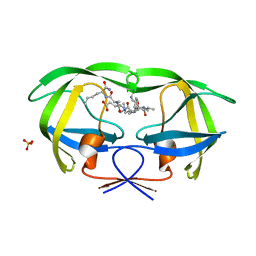 | | Combining Mutations in HIV-1 Protease to Understand Mechanisms of Resistance | | 分子名称: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN, SULFATE ION | | 著者 | Mahalingam, B, Boross, P, Wang, Y.-F, Louis, J.M, Fischer, C, Tozser, J, Harrison, R.W, Weber, I.T. | | 登録日 | 2001-09-25 | | 公開日 | 2002-07-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Combining mutations in HIV-1 protease to understand mechanisms of resistance.
Proteins, 48, 2002
|
|
1K2C
 
 | | Combining Mutations in HIV-1 Protease to Understand Mechanisms of Resistance | | 分子名称: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN | | 著者 | Mahalingam, B, Boross, P, Wang, Y.-F, Louis, J.M, Fischer, C, Tozser, J, W Harrison, R, Weber, I.T. | | 登録日 | 2001-09-26 | | 公開日 | 2002-07-10 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Combining mutations in HIV-1 protease to understand mechanisms of resistance.
Proteins, 48, 2002
|
|
1A8F
 
 | | HUMAN SERUM TRANSFERRIN, RECOMBINANT N-TERMINAL LOBE | | 分子名称: | CARBONATE ION, FE (III) ION, SERUM TRANSFERRIN | | 著者 | Macgillivray, R.T.A, Moore, S.A, Chen, J, Anderson, B.F, Baker, H, Luo, Y, Bewley, M, Smith, C.A, Murphy, M.E.P, Wang, Y, Mason, A.B, Woodworth, R.C, Brayer, G.D, Baker, E.N. | | 登録日 | 1998-03-25 | | 公開日 | 1998-06-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Two high-resolution crystal structures of the recombinant N-lobe of human transferrin reveal a structural change implicated in iron release.
Biochemistry, 37, 1998
|
|
5DGU
 
 | | Crystal Structure of HIV-1 Protease Inhibitors Containing Substituted fused-Tetrahydropyranyl Tetrahydrofuran as P2-Ligand GRL-004-11A | | 分子名称: | (3R,3aR,4S,7aS)-3-methoxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | 著者 | Agniswamy, J, Wang, Y.-F, Weber, I.T. | | 登録日 | 2015-08-28 | | 公開日 | 2015-10-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Design, synthesis, biological evaluation and X-ray structural studies of HIV-1 protease inhibitors containing substituted fused-tetrahydropyranyl tetrahydrofuran as P2-ligands.
Org.Biomol.Chem., 13, 2015
|
|
4J1J
 
 | | Leanyer orthobunyavirus nucleoprotein-ssDNA complex | | 分子名称: | DNA (5'-D(P*AP*CP*CP*AP*AP*AP*CP*AP*AP*CP*CP*CP*AP*CP*CP*CP*A)-3'), Nucleocapsid | | 著者 | Niu, F, Shaw, N, Wang, Y, Jiao, L, Ding, W, Li, X, Zhu, P, Upur, H, Ouyang, S, Cheng, G, Liu, Z.J. | | 登録日 | 2013-02-01 | | 公開日 | 2013-04-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structure of the Leanyer orthobunyavirus nucleoprotein-RNA complex reveals unique architecture for RNA encapsidation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6JEA
 
 | | crystal structure of a beta-N-acetylhexosaminidase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, ZINC ION | | 著者 | Chen, X, Wang, J.C, Liu, M.J, Yang, W.Y, Wang, Y.Z, Tang, R.P, Zhang, M. | | 登録日 | 2019-02-04 | | 公開日 | 2019-03-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.275 Å) | | 主引用文献 | Crystallographic evidence for substrate-assisted catalysis of beta-N-acetylhexosaminidas from Akkermansia muciniphila.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
5YEH
 
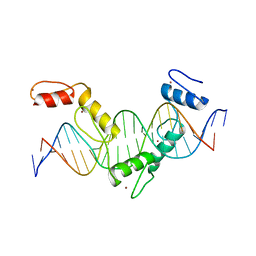 | | Crystal structure of CTCF ZFs4-8-eCBS | | 分子名称: | DNA (5'-D(*AP*CP*GP*GP*TP*TP*TP*CP*CP*GP*CP*TP*AP*GP*AP*GP*GP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*CP*CP*TP*CP*TP*AP*GP*CP*GP*GP*AP*AP*AP*CP*CP*G)-3'), Transcriptional repressor CTCF, ... | | 著者 | Yin, M, Wang, J, Wang, M, Li, X, Wang, Y. | | 登録日 | 2017-09-17 | | 公開日 | 2017-11-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.328 Å) | | 主引用文献 | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
4ZHW
 
 | |
