4DSO
 
 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | 分子名称: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, BENZAMIDINE, GLYCEROL, ... | | 著者 | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | 登録日 | 2012-02-19 | | 公開日 | 2012-04-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DSU
 
 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | 分子名称: | BENZIMIDAZOLE, GTPase KRas, isoform 2B, ... | | 著者 | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | 登録日 | 2012-02-19 | | 公開日 | 2012-04-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5YIM
 
 | |
7MDP
 
 | | KRas G12C in complex with G-2897 | | 分子名称: | 1,2-ETHANEDIOL, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 4-(trifluoromethyl)-1,3-benzothiazol-2-amine, ... | | 著者 | Oh, A, Frank, Y, Wang, W. | | 登録日 | 2021-04-05 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Conformation-locking antibodies for the discovery and characterization of KRAS inhibitors.
Nat.Biotechnol., 40, 2022
|
|
6CQD
 
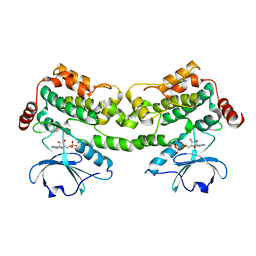 | | Crystal structure of HPK1 in complex with ATP analogue (AMPPNP) | | 分子名称: | MAGNESIUM ION, Mitogen-activated protein kinase kinase kinase kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Wu, P, Lehoux, I, Franke, Y, Mortara, K, Wang, W. | | 登録日 | 2018-03-14 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Hematopoietic Progenitor Kinase-1 Structure in a Domain-Swapped Dimer.
Structure, 27, 2019
|
|
5GXT
 
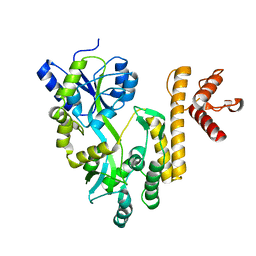 | | Crystal structure of PigG | | 分子名称: | MAGNESIUM ION, Maltose-binding periplasmic protein,PigG | | 著者 | Zhang, F, Ran, T, Xu, D, Wang, W. | | 登録日 | 2016-09-20 | | 公開日 | 2017-07-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.245 Å) | | 主引用文献 | Crystal structure of MBP-PigG fusion protein and the essential function of PigG in the prodigiosin biosynthetic pathway in Serratia marcescens FS14.
Int. J. Biol. Macromol., 99, 2017
|
|
5URY
 
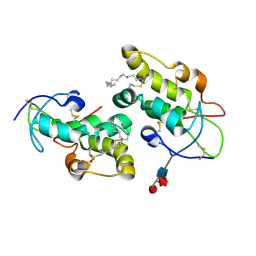 | | Crystal structure of Frizzled 5 CRD in complex with PAM | | 分子名称: | Frizzled-5, PALMITOLEIC ACID, alpha-L-fucopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Mukund, S, Nile, A.H, Stanger, K, Hannoush, R.N, Wang, W. | | 登録日 | 2017-02-13 | | 公開日 | 2017-05-10 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.098 Å) | | 主引用文献 | Unsaturated fatty acyl recognition by Frizzled receptors mediates dimerization upon Wnt ligand binding.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6CQE
 
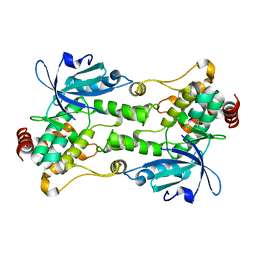 | | Crystal structure of HPK1 kinase domain S171A mutant | | 分子名称: | Mitogen-activated protein kinase kinase kinase kinase 1 | | 著者 | Wu, P, Lehoux, I, Mortara, K, Franke, Y, Wang, W. | | 登録日 | 2018-03-15 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.886 Å) | | 主引用文献 | Hematopoietic Progenitor Kinase-1 Structure in a Domain-Swapped Dimer.
Structure, 27, 2019
|
|
5TUB
 
 | | Crystal Structure of the Shark TBC1D15 GAP Domain | | 分子名称: | Shark TBC1D15 GTPase-activating Protein | | 著者 | Chen, Y.-N, Wang, W, Cheng, D, Ge, Y, Gu, X, Zhou, X.E, Ye, F, Xu, H.E, Lv, Z. | | 登録日 | 2016-11-05 | | 公開日 | 2017-02-22 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Crystal structure of TBC1D15 GTPase-activating protein (GAP) domain and its activity on Rab GTPases.
Protein Sci., 26, 2017
|
|
5TUC
 
 | | Crystal Structure of the Sus TBC1D15 GAP Domain | | 分子名称: | Sus TBC1D15 GAP Domain | | 著者 | Chen, Y.-N, Wang, W, Cheng, D, Ge, Y, Gu, X, Zhou, X.E, Ye, F, Xu, H.E, Lv, Z. | | 登録日 | 2016-11-05 | | 公開日 | 2017-02-22 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of TBC1D15 GTPase-activating protein (GAP) domain and its activity on Rab GTPases.
Protein Sci., 26, 2017
|
|
5GWD
 
 | | Structure of Myroilysin | | 分子名称: | Myroilysin, ZINC ION | | 著者 | Xu, D, Ran, T, Wang, W. | | 登録日 | 2016-09-10 | | 公開日 | 2017-02-15 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Myroilysin Is a New Bacterial Member of the M12A Family of Metzincin Metallopeptidases and Is Activated by a Cysteine Switch Mechanism.
J. Biol. Chem., 292, 2017
|
|
5URV
 
 | | Crystal structure of Frizzled 7 CRD in complex with C24 fatty acid | | 分子名称: | (15E)-TETRACOS-15-ENOIC ACID, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Mukund, S, Nile, A.H, Stanger, K, Hannoush, R.N, Wang, W. | | 登録日 | 2017-02-13 | | 公開日 | 2017-05-10 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Unsaturated fatty acyl recognition by Frizzled receptors mediates dimerization upon Wnt ligand binding.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5URZ
 
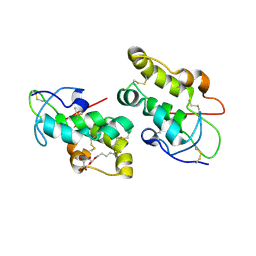 | | Crystal structure of Frizzled 5 CRD in complex with BOG | | 分子名称: | Frizzled-5, alpha-L-fucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, octyl beta-D-glucopyranoside | | 著者 | Mukund, S, Nile, A.H, Stanger, K, Hannoush, R.N, Wang, W. | | 登録日 | 2017-02-13 | | 公開日 | 2017-05-10 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.202 Å) | | 主引用文献 | Unsaturated fatty acyl recognition by Frizzled receptors mediates dimerization upon Wnt ligand binding.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5DPQ
 
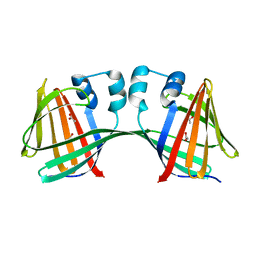 | | Crystal Structure of E72A mutant of domain swapped dimer Human Cellular Retinol Binding Protein | | 分子名称: | ACETATE ION, Retinol-binding protein 2 | | 著者 | Assar, Z, Nossoni, Z, Wang, W, Geiger, J.H, Borhan, B. | | 登録日 | 2015-09-14 | | 公開日 | 2016-09-14 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.775 Å) | | 主引用文献 | Domain-Swapped Dimers of Intracellular Lipid-Binding Proteins: Evidence for Ordered Folding Intermediates.
Structure, 24, 2016
|
|
7UFZ
 
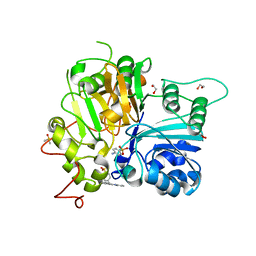 | | Crystal structure of TDP1 complexed with compound XZ768 | | 分子名称: | (4-{[(4S)-2-phenylimidazo[1,2-a]pyridin-3-yl]amino}phenyl)phosphonic acid, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | 著者 | Lountos, G.T, Zhao, X.Z, Wang, W, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R. | | 登録日 | 2022-03-23 | | 公開日 | 2023-04-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.559 Å) | | 主引用文献 | Phosphonic acid-containing inhibitors of tyrosyl-DNA phosphodiesterase 1.
Front Chem, 10, 2022
|
|
7UFY
 
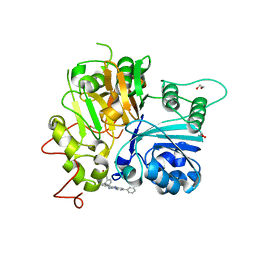 | | Crystal structure of TDP1 complexed with compound XZ766 | | 分子名称: | 1,2-ETHANEDIOL, Tyrosyl-DNA phosphodiesterase 1, [(4-{[(4S)-2,7-diphenylimidazo[1,2-a]pyridin-3-yl]amino}phenyl)methyl]phosphonic acid | | 著者 | Lountos, G.T, Zhao, X.Z, Wang, W, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R. | | 登録日 | 2022-03-23 | | 公開日 | 2023-04-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.584 Å) | | 主引用文献 | Phosphonic acid-containing inhibitors of tyrosyl-DNA phosphodiesterase 1.
Front Chem, 10, 2022
|
|
5FGL
 
 | | Co-crystal Structure of NicR2_Hsp | | 分子名称: | 4-oxidanylidene-4-(6-oxidanylidene-1~{H}-pyridin-3-yl)butanoic acid, NicR | | 著者 | Zhang, K, Tang, H, Wu, G, Wang, W, Hu, H, Xu, P. | | 登録日 | 2015-12-21 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Co-crystal Structure of NicR2_Hsp
To Be Published
|
|
5GXV
 
 | | Crystal structure of PigG | | 分子名称: | MAGNESIUM ION, Maltose-binding periplasmic protein,PigG | | 著者 | Zhang, F, Ran, T, Xu, D, Wang, W. | | 登録日 | 2016-09-20 | | 公開日 | 2017-07-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of MBP-PigG fusion protein and the essential function of PigG in the prodigiosin biosynthetic pathway in Serratia marcescens FS14.
Int. J. Biol. Macromol., 99, 2017
|
|
6CQF
 
 | | Crystal structure of HPK1 in complex an inhibitor G1858 | | 分子名称: | Mitogen-activated protein kinase kinase kinase kinase 1, N-{2-(3,3-difluoropyrrolidin-1-yl)-6-[(3R)-pyrrolidin-3-yl]pyrimidin-4-yl}-1-(propan-2-yl)-1H-pyrazolo[4,3-c]pyridin-6-amine | | 著者 | Wu, P, Lehoux, I, Mortara, K, Franke, Y, Chan, B.K, Wang, W. | | 登録日 | 2018-03-15 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.246 Å) | | 主引用文献 | Hematopoietic Progenitor Kinase-1 Structure in a Domain-Swapped Dimer.
Structure, 27, 2019
|
|
4DST
 
 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | 分子名称: | 1,2-ETHANEDIOL, 2-(4,6-dichloro-2-methyl-1H-indol-3-yl)ethanamine, ACETATE ION, ... | | 著者 | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | 登録日 | 2012-02-19 | | 公開日 | 2012-04-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DSN
 
 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | 分子名称: | 1,2-ETHANEDIOL, GTPase KRas, isoform 2B, ... | | 著者 | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | 登録日 | 2012-02-19 | | 公開日 | 2012-04-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3LF5
 
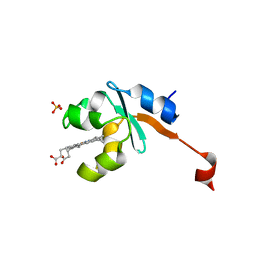 | | Structure of Human NADH cytochrome b5 oxidoreductase (Ncb5or) b5 Domain to 1.25A Resolution | | 分子名称: | Cytochrome b5 reductase 4, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | 著者 | Deng, B, Parthasarathy, S, Wang, W, Gibney, B.R, Battaile, K.P, Lovell, S, Benson, D.R, Zhu, H. | | 登録日 | 2010-01-15 | | 公開日 | 2010-07-14 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Study of the individual cytochrome b5 and cytochrome b5 reductase domains of Ncb5or reveals a unique heme pocket and a possible role of the CS domain.
J.Biol.Chem., 285, 2010
|
|
6D5Y
 
 | |
5H9U
 
 | |
3L82
 
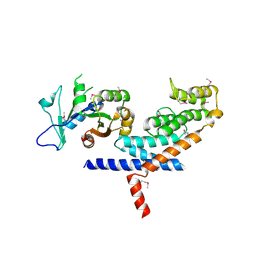 | | X-ray Crystal structure of TRF1 and Fbx4 complex | | 分子名称: | F-box only protein 4, Telomeric repeat-binding factor 1 | | 著者 | Zeng, Z.X, Wang, W, Yang, Y.T, Chen, Y, Yang, X.M, Diehl, J.A, Liu, X.D, Lei, M. | | 登録日 | 2009-12-29 | | 公開日 | 2010-03-09 | | 最終更新日 | 2013-09-25 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Basis of Selective Ubiquitination of TRF1 by SCF(Fbx4)
Dev.Cell, 18, 2010
|
|
