5LIN
 
 | |
5LIS
 
 | |
2QDY
 
 | | Crystal Structure of Fe-type NHase from Rhodococcus erythropolis AJ270 | | Descriptor: | 2-METHYLPROPAN-1-AMINE, CHLORIDE ION, FE (III) ION, ... | | Authors: | Song, L, Shi, J, Xue, Z, Wang, M.-X, Qian, S. | | Deposit date: | 2007-06-22 | | Release date: | 2008-05-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High resolution X-ray molecular structure of the nitrile hydratase from Rhodococcus erythropolis AJ270 reveals posttranslational oxidation of two cysteines into sulfinic acids and a novel biocatalytic nitrile hydration mechanism
Biochem.Biophys.Res.Commun., 362, 2007
|
|
8Q2P
 
 | | Structure of the membrane integral lipoprotein N-acyltransferase Lnt from E. coli by using Se-MAG for the the lipid cubic phase crystallization | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Apolipoprotein N-acyltransferase, ... | | Authors: | Huang, C.-Y, Boland, C, Kaki, S.S, Wang, M, Olieric, V, Caffrey, M. | | Deposit date: | 2023-08-03 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Se-MAG Is a Convenient Additive for Experimental Phasing and Structure Determination of Membrane Proteins Crystallised by the Lipid Cubic Phase (In Meso) Method
Crystals, 2023
|
|
8AQ2
 
 | | In meso structure of the membrane integral lipoprotein N-acyltransferase Lnt from P. aeruginosa covalently linked with TITC | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, CITRATE ANION, ... | | Authors: | Huang, C.-Y, Weichert, D, Boland, C, Smithers, L, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2022-08-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure snapshots reveal the mechanism of a bacterial membrane lipoprotein N -acyltransferase.
Sci Adv, 9, 2023
|
|
4KUD
 
 | | Crystal structure of N-terminal acetylated Sir3 BAH domain D205N mutant in complex with yeast nucleosome core particle | | Descriptor: | Histone H2A.2, Histone H2B.1, Histone H3, ... | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.203 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
1XXO
 
 | | X-ray crystal structure of mycobacterium tuberculosis pyridoxine 5'-phosphate oxidase at 1.8 a resolution | | Descriptor: | hypothetical protein Rv1155 | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Garen, C, James, M.N.G, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-11-07 | | Release date: | 2004-11-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of mycobacterium tuberculosis pyridoxine 5'-phosphate oxidase at 1.8 a resolution
To be Published
|
|
6KY4
 
 | | Crystal structure of Sulfiredoxin from Arabidopsis thaliana | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, Sulfiredoxin, ... | | Authors: | Liu, M, Wang, J, Li, X, Li, M, Sylvanno, M.J, Zhang, M, Wang, M. | | Deposit date: | 2019-09-16 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of sulfiredoxin from Arabidopsis thaliana revealed a more robust antioxidant mechanism in plants.
Biochem.Biophys.Res.Commun., 520, 2019
|
|
1Y30
 
 | | X-ray crystal structure of mycobacterium tuberculosis pyridoxine 5'-phosphate oxidase complexed with flavin mononucleotide at 2.2 a resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, hypothetical protein Rv1155 | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Garen, C, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-11-23 | | Release date: | 2005-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Mycobacterium tuberculosispyridoxine 5'-phosphate oxidase and its complexes with flavin mononucleotide and pyridoxal 5'-phosphate.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3ZJ6
 
 | | Crystal of Raucaffricine Glucosidase in complex with inhibitor | | Descriptor: | (1R,2S,3S,4R,5R)-4-(cyclohexylmethylamino)-5-(hydroxymethyl)cyclopentane-1,2,3-triol, RAUCAFFRICINE-O-BETA-D-GLUCOSIDASE, SULFATE ION | | Authors: | Xia, L, Lin, H, Panjikar, S, Ruppert, M, Castiglia, A, Rajendran, C, Wang, M, Schuebel, H, Warzecha, H, Jaeger, V, Stoeckigt, J. | | Deposit date: | 2013-01-17 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ligand Structures of Synthetic Deoxa-Pyranosylamines with Raucaffricine and Strictosidine Glucosidases Provide Structural Insights Into Their Binding and Inhibitory Behaviours.
J.Enzyme.Inhib.Med.Chem., 30, 2015
|
|
1KV0
 
 | | Cis/trans Isomerization of Non-prolyl Peptide Bond Observed in Crystal Structure of an Scorpion Toxin | | Descriptor: | Alpha-like toxin BmK-M7 | | Authors: | Guan, R.J, He, X.L, Wang, M, Xiang, Y, Wang, D.C. | | Deposit date: | 2002-01-23 | | Release date: | 2003-09-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural mechanism governing cis and trans isomeric states and an intramolecular switch for cis/trans isomerization of a non-proline peptide bond observed in crystal structures of scorpion toxins.
J.Mol.Biol., 341, 2004
|
|
6B3J
 
 | | 3.3 angstrom phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex | | Descriptor: | Exendin-P5, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liang, Y.L, Khoshouei, M, Glukhova, A, Furness, S.G.B, Koole, C, Zhao, P, Clydesdale, L, Thal, D.M, Radjainia, M, Danev, R, Baumeister, W, Wang, M.W, Miller, L.J, Christopoulos, A, Sexton, P.M, Wootten, D. | | Deposit date: | 2017-09-22 | | Release date: | 2018-02-21 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex.
Nature, 555, 2018
|
|
1YVX
 
 | | Hepatitis C Virus RNA Polymerase Genotype 2a In Complex With Non- Nucleoside Analogue Inhibitor | | Descriptor: | 3-[ISOPROPYL(4-METHYLBENZOYL)AMINO]-5-PHENYLTHIOPHENE-2-CARBOXYLIC ACID, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the RNA dependent RNA polymerase genotype 2a of hepatitis C virus reveal two conformations and suggest mechanisms of inhibition by non-nucleoside inhibitors.
J.Biol.Chem., 280, 2005
|
|
5D58
 
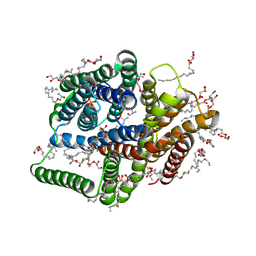 | | In meso in situ serial X-ray crystallography structure of the PepTSt-Ala-Phe complex at 100 K | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ALANINE, ... | | Authors: | Huang, C.-Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5JWY
 
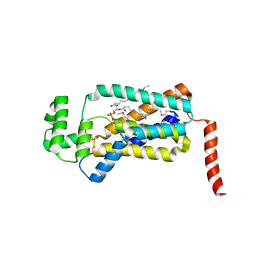 | | Structure of lipid phosphate phosphatase PgpB complex with PE | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate, Phosphatidylglycerophosphatase B | | Authors: | Tong, S, Wang, M, Zheng, L. | | Deposit date: | 2016-05-12 | | Release date: | 2016-07-20 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Insight into Substrate Selection and Catalysis of Lipid Phosphate Phosphatase PgpB in the Cell Membrane.
J.Biol.Chem., 291, 2016
|
|
8GQC
 
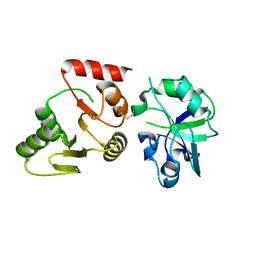 | | Crystal structure of the SARS-unique domain (SUD) of SARS-CoV-2 (1.35 angstrom resolution) | | Descriptor: | Papain-like protease nsp3 | | Authors: | Qin, B, Li, Z, Aumonier, S, Wang, M, Cui, S. | | Deposit date: | 2022-08-30 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Identification of the SARS-unique domain of SARS-CoV-2 as an antiviral target.
Nat Commun, 14, 2023
|
|
7Y5K
 
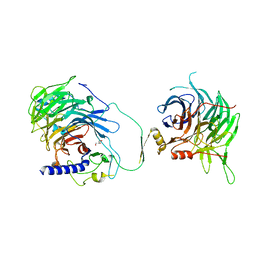 | | Crystal structure of human CAF-1 core complex in spacegroup C2221 | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, GLYCEROL, ... | | Authors: | Liu, C.P, Wang, M.Z, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
5ED9
 
 | | Crystal structure of CC1 of mouse SUN2 | | Descriptor: | SUN domain-containing protein 2 | | Authors: | Nie, S, Ke, H.M, Gao, F, Ren, J.Q, Wang, M.Z, Huo, L, Gong, W.M, Feng, W. | | Deposit date: | 2015-10-21 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Coiled-Coil Domains of SUN Proteins as Intrinsic Dynamic Regulators
Structure, 24, 2016
|
|
5ZBA
 
 | | Crystal structure of Rtt109-Asf1-H3-H4-CoA complex | | Descriptor: | COENZYME A, DNA damage response protein Rtt109, putative, ... | | Authors: | Zhang, L, Serra-Cardona, A, Zhou, H, Wang, M, Yang, N, Zhang, Z, Xu, R.M. | | Deposit date: | 2018-02-10 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Multisite Substrate Recognition in Asf1-Dependent Acetylation of Histone H3 K56 by Rtt109.
Cell, 174, 2018
|
|
5E5A
 
 | | Crystal structure of the chromatin-tethering domain of Human cytomegalovirus IE1 protein bound to the nucleosome core particle | | Descriptor: | C-terminal domain of Regulatory protein IE1, DNA (146-MER), Histone H2A, ... | | Authors: | Fang, Q, Chen, P, Wang, M, Fang, J, Yang, N, Li, G, Xu, R.M. | | Deposit date: | 2015-10-08 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Human cytomegalovirus IE1 protein alters the higher-order chromatin structure by targeting the acidic patch of the nucleosome
Elife, 5, 2016
|
|
5VEX
 
 | | Structure of the human GLP-1 receptor complex with NNC0640 | | Descriptor: | 4-{[(4-cyclohexylphenyl){[3-(methylsulfonyl)phenyl]carbamoyl}amino]methyl}-N-(1H-tetrazol-5-yl)benzamide, Glucagon-like peptide 1 receptor, Endolysin chimera | | Authors: | Song, G, Yang, D, Wang, Y, Graaf, C.D, Zhou, Q, Jiang, S, Liu, K, Cai, X, Dai, A, Lin, G, Liu, D, Wu, F, Wu, Y, Zhao, S, Ye, L, Han, G.W, Lau, J, Wu, B, Hanson, M.A, Liu, Z.-J, Wang, M.-W, Stevens, R.C. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators.
Nature, 546, 2017
|
|
5ZBB
 
 | | Crystal structure of Rtt109-Asf1-H3-H4 complex | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA damage response protein Rtt109, putative, ... | | Authors: | Zhang, L, Serra-Cardona, A, Zhou, H, Wang, M, Yang, N, Zhang, Z, Xu, R.M. | | Deposit date: | 2018-02-10 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Multisite Substrate Recognition in Asf1-Dependent Acetylation of Histone H3 K56 by Rtt109.
Cell, 174, 2018
|
|
8HBL
 
 | | Crystal structure of the SARS-unique domain (SUD) of SARS-CoV-2 (1.58 angstrom resolution) | | Descriptor: | GLYCEROL, LITHIUM ION, Non-structural protein 3, ... | | Authors: | Qin, B, Li, Z, Aumonier, S, Wang, M, Cui, S. | | Deposit date: | 2022-10-29 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Identification of the SARS-unique domain of SARS-CoV-2 as an antiviral target.
Nat Commun, 14, 2023
|
|
6G89
 
 | | Thaumatin solved by Native SAD from a dataset collected in 0.6 second with JUNGFRAU detector | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Leonarski, F, Olieric, V, Vera, L, Redford, S, Wang, M. | | Deposit date: | 2018-04-08 | | Release date: | 2018-08-01 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | Fast and accurate data collection for macromolecular crystallography using the JUNGFRAU detector.
Nat. Methods, 15, 2018
|
|
4BPM
 
 | | Crystal structure of a human integral membrane enzyme | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-[(2,2-dimethylpropanoylamino)methyl]phenyl]amino]-1-methyl-6-(2-methyl-2-oxidanyl-propoxy)-N-[2,2,2-tris(fluoranyl)ethyl]benzimidazole-5-carboxamide, GLUTATHIONE, PROSTAGLANDIN E SYNTHASE, ... | | Authors: | Li, D, Wang, M, Olieric, V, Caffrey, M. | | Deposit date: | 2013-05-27 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystallizing Membrane Proteins in the Lipidic Mesophase. Experience with Human Prostaglandin E2 Synthase 1 and an Evolving Strategy.
Cryst.Growth Des., 14, 2014
|
|
