7X01
 
 | | Cryo-EM Structure of Chikungunya Virus Nonstructural Protein 1 with inhibitor FHA | | Descriptor: | (1R,2S,3S,4R,5R)-3-(6-aminopurin-9-yl)-4-fluoranyl-5-(2-hydroxyethyl)cyclopentane-1,2-diol, ZINC ION, mRNA-capping enzyme nsP1 | | Authors: | Zhang, K, Law, M.C.Y, Nguyen, T.M, Tan, Y.B, Wirawan, M, Law, Y.S, Luo, D.H. | | Deposit date: | 2022-02-20 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Molecular basis of specific viral RNA recognition and 5'-end capping by the Chikungunya virus nsP1.
Cell Rep, 40, 2022
|
|
7Y5K
 
 | | Crystal structure of human CAF-1 core complex in spacegroup C2221 | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, GLYCEROL, ... | | Authors: | Liu, C.P, Wang, M.Z, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
5YPS
 
 | | The structural basis of histone chaperoneVps75 | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Chen, Y, Zhang, Y, Dou, Y, Wang, M, Xu, S, Jiang, H, Limper, A, Su, D. | | Deposit date: | 2017-11-03 | | Release date: | 2018-11-07 | | Last modified: | 2020-06-10 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structural basis for the acetylation of histone H3K9 and H3K27 mediated by the histone chaperone Vps75 inPneumocystis carinii.
Signal Transduct Target Ther, 4, 2019
|
|
2MLB
 
 | | NMR solution structure of a computational designed protein based on template of human erythrocytic ubiquitin | | Descriptor: | redesigned ubiquitin | | Authors: | Xiong, P, Wang, M, Zhang, J, Chen, Q, Liu, H. | | Deposit date: | 2014-02-21 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein design with a comprehensive statistical energy function and boosted by experimental selection for foldability
Nat Commun, 5, 2014
|
|
2MN4
 
 | | NMR solution structure of a computational designed protein based on structure template 1cy5 | | Descriptor: | Computational designed protein based on structure template 1cy5 | | Authors: | Xiong, P, Wang, M, Zhang, J, Chen, Q, Liu, H. | | Deposit date: | 2014-03-28 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein design with a comprehensive statistical energy function and boosted by experimental selection for foldability
Nat Commun, 5, 2014
|
|
3QNO
 
 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dATP Opposite 3tCo | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA Polymerase, ... | | Authors: | Xia, S, Wang, M, Wang, J, Konigsberg, W.H. | | Deposit date: | 2011-02-08 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Using a Fluorescent Cytosine Analogue tC(o) To Probe the Effect of the Y567 to Ala Substitution on the Preinsertion Steps of dNMP Incorporation by RB69 DNA Polymerase.
Biochemistry, 51, 2012
|
|
2RSG
 
 | | Solution structure of the CERT PH domain | | Descriptor: | Collagen type IV alpha-3-binding protein | | Authors: | Sugiki, T, Takeuchi, K, Tokunaga, Y, Kumagai, K, Kawano, M, Nishijima, M, Hanada, K, Takahashi, H, Shimada, I. | | Deposit date: | 2012-02-25 | | Release date: | 2012-08-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the Golgi association by the pleckstrin homology domain of the ceramide trafficking protein (CERT)
J.Biol.Chem., 287, 2012
|
|
7WIT
 
 | | Structure of SUR1 in complex with mitiglinide | | Descriptor: | (2S)-4-[(3aR,7aS)-1,3,3a,4,5,6,7,7a-octahydroisoindol-2-yl]-4-oxidanylidene-2-(phenylmethyl)butanoic acid, ADENOSINE-5'-TRIPHOSPHATE, ATP-sensitive inward rectifier potassium channel 11,ATP-binding cassette sub-family C member 8 isoform X1 | | Authors: | Chen, L, Wang, M.M. | | Deposit date: | 2022-01-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural Insights Into the High Selectivity of the Anti-Diabetic Drug Mitiglinide
Front Pharmacol, 13, 2022
|
|
4GXL
 
 | | The crystal structure of Galectin-8 C-CRD in complex with NDP52 | | Descriptor: | GLYCEROL, Galectin-8, Peptide from Calcium-binding and coiled-coil domain-containing protein 2 | | Authors: | Li, S, Wandel, M.P, Li, F, Liu, Z, He, C, Wu, J, Shi, Y, Randow, F. | | Deposit date: | 2012-09-04 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.023 Å) | | Cite: | Sterical hindrance promotes selectivity of the autophagy cargo receptor NDP52 for the danger receptor galectin-8 in antibacterial autophagy
Sci.Signal., 6, 2013
|
|
7YK6
 
 | | Cryo-EM structure of the compound 4-bound human relaxin family peptide receptor 4 (RXFP4)-Gi complex | | Descriptor: | 1-[2-(4-chlorophenyl)ethyl]-3-[(7-ethyl-5-oxidanyl-1H-indol-3-yl)methylideneamino]guanidine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Chen, Y, Zhou, Q.T, Wang, J, Xu, Y.W, Wang, Y, Yan, J.H, Wang, Y.B, Zhu, Q, Zhao, F.H, Li, C.H, Chen, C.W, Cai, X.Q, Bathgate, R.A.D, Shen, C, Liu, H, Xu, H.E, Yang, D.H, Wang, M.W. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Ligand recognition mechanism of the human relaxin family peptide receptor 4 (RXFP4).
Nat Commun, 14, 2023
|
|
7YJ4
 
 | | Cryo-EM structure of the INSL5-bound human relaxin family peptidereceptor 4 (RXFP4)-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-2, ... | | Authors: | Chen, Y, Zhou, Q.T, Wang, J, Xu, Y.W, Wang, Y, Yan, J.H, Wang, Y.B, Zhu, Q, Zhao, F.H, Li, C.H, Chen, C.W, Cai, X.Q, Bathgate, R.A.D, Shen, C, Liu, H, Xu, H.E, Yang, D.H, Wang, M.W. | | Deposit date: | 2022-07-19 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Ligand recognition mechanism of the human relaxin family peptide receptor 4 (RXFP4).
Nat Commun, 14, 2023
|
|
7YK7
 
 | | Cryo-EM structure of the DC591053-bound human relaxin family peptide receptor 4 (RXFP4)-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-2, ... | | Authors: | Chen, Y, Zhou, Q.T, Wang, J, Xu, Y.W, Wang, Y, Yan, J.H, Wang, Y.B, Zhu, Q, Zhao, F.H, Li, C.H, Chen, C.W, Cai, X.Q, Bathgate, R.A.D, Shen, C, Xu, H.E, Yang, D.H, Liu, H, Wang, M.W. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Ligand recognition mechanism of the human relaxin family peptide receptor 4 (RXFP4).
Nat Commun, 14, 2023
|
|
5XM2
 
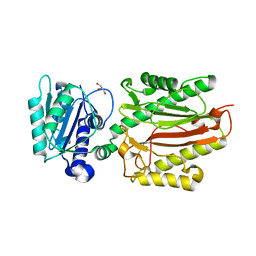 | | Human N-terminal domain of FACT complex subunit SPT16 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FACT complex subunit SPT16, GLYCEROL | | Authors: | Xu, S, Li, H, Dou, Y, Chen, Y, Jiang, H, Lu, D, Wang, M, Su, D. | | Deposit date: | 2017-05-12 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | The structural basis of human Spt16 N-terminal domain interaction with histone (H3-H4)2tetramer.
Biochem.Biophys.Res.Commun., 508, 2019
|
|
7W4P
 
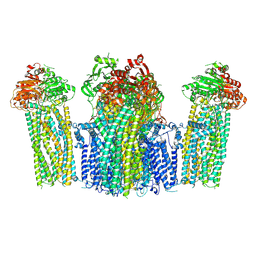 | | The structure of KATP H175K mutant in closed state | | Descriptor: | 6-chloranyl-~{N}-(1-methylcyclopropyl)-1,1-bis(oxidanylidene)-4~{H}-thieno[3,2-e][1,2,4]thiadiazin-3-amine, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, L, Wang, M. | | Deposit date: | 2021-11-28 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural insights into the mechanism of pancreatic K ATP channel regulation by nucleotides.
Nat Commun, 13, 2022
|
|
7W4O
 
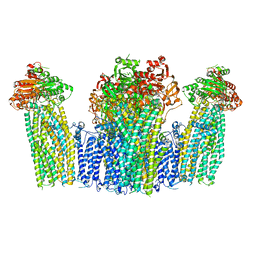 | | The structure of KATP H175K mutant in pre-open state | | Descriptor: | 6-chloranyl-~{N}-(1-methylcyclopropyl)-1,1-bis(oxidanylidene)-4~{H}-thieno[3,2-e][1,2,4]thiadiazin-3-amine, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, L, Wang, M. | | Deposit date: | 2021-11-28 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural insights into the mechanism of pancreatic K ATP channel regulation by nucleotides.
Nat Commun, 13, 2022
|
|
2DHS
 
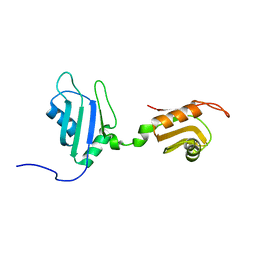 | | Solution Structure of Nucleic Acid Binding Protein CUGBP1ab and its Binding Study with DNA and RNA | | Descriptor: | CUG triplet repeat RNA-binding protein 1 | | Authors: | Xia, Y.L, Jun, K.Y, Zhu, Q, Han, X.G, Zhang, H, Timchenko, L, Swanson, M, Gao, X.L. | | Deposit date: | 2006-03-25 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Nucleic Acid Binding Protein CUGBP1ab and its Binding Study with DNA and RNA
To be Published
|
|
3HM1
 
 | | Crystal structure of human Estrogen Receptor Alpha Ligand-Binding Domain in complex with a Glucocorticoid Receptor Interacting Protein 1 Nr Box II Peptide and estrone ((8R,9S,13S,14S)-3-hydroxy-13-methyl-7,8,9,11,12,14,15,16-octahydro-6H-cyclopenta[a]phenanthren-17-one) | | Descriptor: | (9beta,13alpha)-3-hydroxyestra-1,3,5(10)-trien-17-one, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Rajan, S.S, Kim, Y, Vanek, K, Liwanag, M, Joachimiak, A, Greene, G.L. | | Deposit date: | 2009-05-28 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of human Estrogen Receptor Alpha Ligand-Binding Domain in complex with a Glucocorticoid Receptor Interacting Protein 1 Nr Box II Peptide and estrone ((8R,9S,13S,14S)-3-hydroxy-13-methyl-7,8,9,11,12,14,15, 16-octahydro-6H-cyclopenta[a]phenanthren-17-one)
To be Published
|
|
2E66
 
 | | Crystal Structure Of CutA1 From Pyrococcus Horikoshii OT3, Mutation D60A | | Descriptor: | CHLORIDE ION, Divalent-cation tolerance protein cutA, SODIUM ION | | Authors: | Bagautdinov, B, Sawano, M, Bagautdinova, S, Yutani, K, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-25 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the hyper-thermostability of CutA1
To be Published
|
|
7X8R
 
 | | Cryo-EM structure of the Boc5-bound hGLP-1R-Gs complex | | Descriptor: | 2,4-bis(3-methoxy-4-thiophen-2-ylcarbonyloxy-phenyl)-1,3-bis[[4-[(2-methylpropan-2-yl)oxycarbonylamino]phenyl]carbonylamino]cyclobutane-1,3-dicarboxylic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z.T, Zhou, Q.T, Li, Y, Chen, L.N, Zhang, Z.C, Liang, A.Y, Liu, Q, Wu, X.Y, Dai, A.T, Xia, T, Wu, W, Zhang, Y, Yang, D.H, Wang, M.W. | | Deposit date: | 2022-03-14 | | Release date: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural basis of peptidomimetic agonism revealed by small- molecule GLP-1R agonists Boc5 and WB4-24.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7X8S
 
 | | Cryo-EM structure of the WB4-24-bound hGLP-1R-Gs complex | | Descriptor: | 2,4-bis(3-methoxy-4-thiophen-2-ylcarbonyloxy-phenyl)-1,3-bis[[4-(2-methylpropanoylamino)phenyl]carbonylamino]cyclobutane-1,3-dicarboxylic acid, Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Cong, Z.T, Zhou, Q.T, Li, Y, Chen, L.N, Zhang, Z.C, Liang, A.Y, Liu, Q, Wu, X.Y, Dai, A.T, Xia, T, Wu, W, Zhang, Y, Yang, D.H, Wang, M.W. | | Deposit date: | 2022-03-14 | | Release date: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis of peptidomimetic agonism revealed by small- molecule GLP-1R agonists Boc5 and WB4-24.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YJT
 
 | |
7WBJ
 
 | | Cryo-EM structure of N-terminal modified human vasoactive intestinal polypeptide receptor 2 (VIP2R) in complex with PACAP27 and Gs | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Xu, Y.N, Feng, W.B, Zhou, Q.T, Liang, A.Y, Li, J, Dai, A.T, Zhao, F.H, Yan, J.H, Chen, C.W, Li, H, Zhao, L.H, Xia, T, Jiang, Y, Xu, H.E, Yang, D.H, Wang, M.W. | | Deposit date: | 2021-12-16 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | A distinctive ligand recognition mechanism by the human vasoactive intestinal polypeptide receptor 2.
Nat Commun, 13, 2022
|
|
7YTW
 
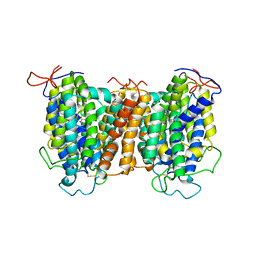 | | Structural basis of vitamin C recognition and transport by mammalian SVCT1 transporter | | Descriptor: | ASCORBIC ACID, SODIUM ION, Solute carrier family 23 member 1 | | Authors: | She, J, Wang, M, He, J, Zhang, K, Li, S. | | Deposit date: | 2022-08-16 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of vitamin C recognition and transport by mammalian SVCT1 transporter.
Nat Commun, 14, 2023
|
|
7YTY
 
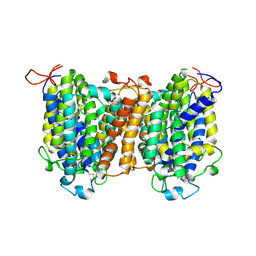 | | Mouse SVCT1 in an apo state | | Descriptor: | Solute carrier family 23 member 1 | | Authors: | She, J, Wang, M, He, J, Zhang, K, Li, S. | | Deposit date: | 2022-08-16 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of vitamin C recognition and transport by mammalian SVCT1 transporter.
Nat Commun, 14, 2023
|
|
4EC6
 
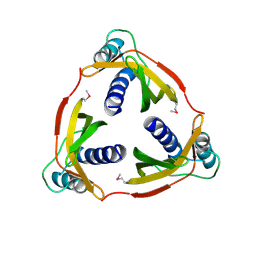 | | Ntf2-like, potential transfer protein TraM from Gram-positive conjugative plasmid pIP501 | | Descriptor: | Putative uncharacterized protein | | Authors: | Goessweiner-Mohr, N, Grumet, L, Pavkov-Keller, T, Wang, M, Keller, W. | | Deposit date: | 2012-03-26 | | Release date: | 2012-12-05 | | Last modified: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 2.5 A Structure of the Enterococcus Conjugation Protein TraM resembles VirB8 Type IV Secretion Proteins.
J.Biol.Chem., 288, 2013
|
|
