6M6K
 
 | |
5JMV
 
 | | Crystal structure of mjKae1-pfuPcc1 complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Probable bifunctional tRNA threonylcarbamoyladenosine biosynthesis protein, ... | | Authors: | Wan, L, Sicheri, F. | | Deposit date: | 2016-04-29 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3864696 Å) | | Cite: | Structural and functional characterization of KEOPS dimerization by Pcc1 and its role in t6A biosynthesis.
Nucleic Acids Res., 44, 2016
|
|
6C80
 
 | | Crystal structure of a flax cytokinin oxidase | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, AMMONIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wan, L, Williams, S, Kobe, B. | | Deposit date: | 2018-01-23 | | Release date: | 2018-06-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and functional insights into the modulation of the activity of a flax cytokinin oxidase by flax rust effector AvrL567-A.
Mol. Plant Pathol., 20, 2019
|
|
7YF7
 
 | |
6M6J
 
 | |
4C6S
 
 | | Crystal structure of the TIR domain from the Arabidopsis Thaliana disease resistance protein RRS1 | | Descriptor: | GLYCEROL, PROBABLE WRKY TRANSCRIPTION FACTOR 52, SODIUM ION, ... | | Authors: | Wan, L, Williams, S.J, Sohn, K.H, Bernoux, M, Ma, Y, Segonzac, C, Ve, T, Sarris, P, Ericsson, D.J, Saucet, S.B, Zhang, X, Parker, J, Dodds, P.N, Jones, J.D.G, Kobe, B. | | Deposit date: | 2013-09-19 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Structural Basis for Assembly and Function of a Heterodimeric Plant Immune Receptor.
Science, 344, 2014
|
|
5J26
 
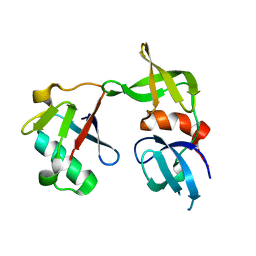 | | Crystal structure of a 53BP1 Tudor domain in complex with a ubiquitin variant | | Descriptor: | Tumor suppressor p53-binding protein 1, Ubiquitin Variant i53 | | Authors: | Wan, L, Canny, M, Juang, Y.C, Durocher, D, Sicheri, F. | | Deposit date: | 2016-03-29 | | Release date: | 2016-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5047 Å) | | Cite: | A genetically encoded inhibitor of 53BP1 to stimulate homology-based gene editing
To Be Published
|
|
7E4E
 
 | |
7D0X
 
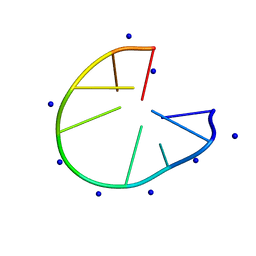 | |
7D0Y
 
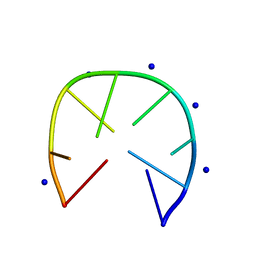 | |
8TQO
 
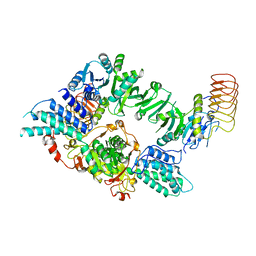 | | Eukaryotic translation initiation factor 2B tetramer | | Descriptor: | Translation initiation factor eIF-2B subunit beta, Translation initiation factor eIF-2B subunit delta, Translation initiation factor eIF-2B subunit epsilon, ... | | Authors: | Wang, L, Lawrence, R, Sangwan, S, Anand, A, Shoemaker, S, Deal, A, Marqusee, S, Watler, P. | | Deposit date: | 2023-08-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A helical fulcrum in eIF2B coordinates allosteric regulation of stress signaling.
Nat.Chem.Biol., 20, 2024
|
|
8TQZ
 
 | | Eukaryotic translation initiation factor 2B with a mutation (L516A) in the delta subunit | | Descriptor: | Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, Translation initiation factor eIF-2B subunit delta, ... | | Authors: | Wang, L, Lawrence, R, Sangwan, S, Anand, A, Shoemaker, S, Deal, A, Marqusee, S, Watler, P. | | Deposit date: | 2023-08-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A helical fulcrum in eIF2B coordinates allosteric regulation of stress signaling.
Nat.Chem.Biol., 20, 2024
|
|
6LNI
 
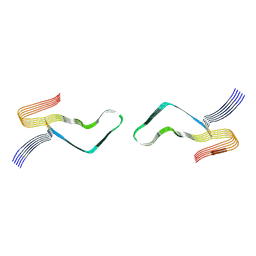 | | Cryo-EM structure of amyloid fibril formed by full-length human prion protein | | Descriptor: | Major prion protein | | Authors: | Wang, L.Q, Zhao, K, Yuan, H.Y, Wang, Q, Guan, Z.Y, Tao, J, Li, X.N, Hao, M.M, Chen, J, Zhang, D.L, Zhu, H.L, Yin, P, Liu, C, Liang, Y. | | Deposit date: | 2019-12-30 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.702 Å) | | Cite: | Cryo-EM structure of an amyloid fibril formed by full-length human prion protein.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7XJA
 
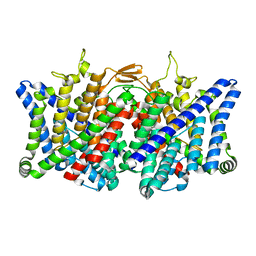 | | TMD masked refine map of human ClC-2 | | Descriptor: | Chloride channel protein 2 | | Authors: | Wang, L. | | Deposit date: | 2022-04-15 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of ClC-2 chloride channel reveal the blocking mechanism of its specific inhibitor AK-42
Nat Commun, 14, 2023
|
|
7XF5
 
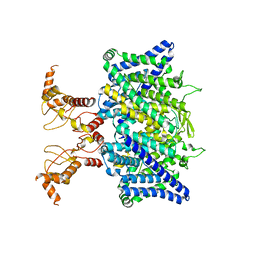 | | Full length human CLC-2 channel in apo state | | Descriptor: | Chloride channel protein 2 | | Authors: | Wang, L. | | Deposit date: | 2022-04-01 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of ClC-2 chloride channel reveal the blocking mechanism of its specific inhibitor AK-42
Nat Commun, 14, 2023
|
|
7RLO
 
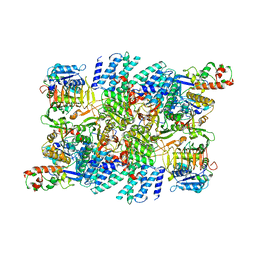 | | Structure of the human eukaryotic translation initiation factor 2B (eIF2B) in complex with a viral protein NSs | | Descriptor: | Non-structural protein NS-S, Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, ... | | Authors: | Wang, L, Schoof, M, Cogan, J, Lawrence, R, Boone, M, Wuerth, J, Frost, M, Walter, P. | | Deposit date: | 2021-07-26 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Viral evasion of the integrated stress response through antagonism of eIF2-P binding to eIF2B.
Nat Commun, 12, 2021
|
|
7RR4
 
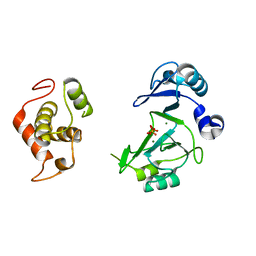 | |
7RR3
 
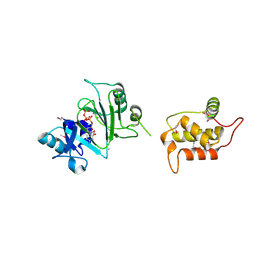 | | Structure of Deep-Sea Phage NrS-1 Primase-Polymerase N300 in complex with calcium and ddCTP | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, CALCIUM ION, Primase | | Authors: | Wang, L, Yu, C, Sliz, P. | | Deposit date: | 2021-08-09 | | Release date: | 2022-01-12 | | Last modified: | 2022-01-19 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Molecular Dissection of the Primase and Polymerase Activities of Deep-Sea Phage NrS-1 Primase-Polymerase.
Front Microbiol, 12, 2021
|
|
6EG9
 
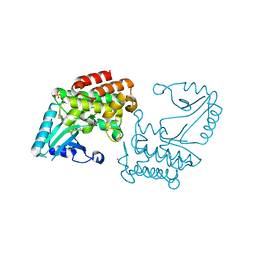 | | IRAK4 in complex with Ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, COBALT (II) ION, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Wang, L, Wu, H. | | Deposit date: | 2018-08-19 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.414 Å) | | Cite: | Conformational flexibility and inhibitor binding to unphosphorylated interleukin-1 receptor-associated kinase 4 (IRAK4).
J.Biol.Chem., 294, 2019
|
|
4X67
 
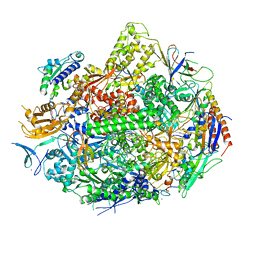 | | Crystal structure of elongating yeast RNA polymerase II stalled at oxidative Cyclopurine DNA lesions. | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2014-12-07 | | Release date: | 2015-02-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Mechanism of RNA polymerase II bypass of oxidative cyclopurine DNA lesions.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6LR0
 
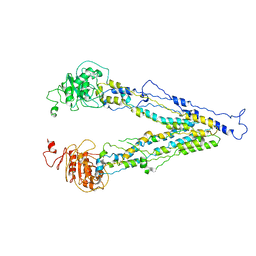 | | structure of human bile salt exporter ABCB11 | | Descriptor: | Bile salt export pump | | Authors: | Wang, L, Hou, W.T, Chen, L, Jiang, Y.L, Xu, D, Sun, L.F, Zhou, C.Z, Chen, Y. | | Deposit date: | 2020-01-15 | | Release date: | 2020-04-15 | | Last modified: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of human bile salts exporter ABCB11.
Cell Res., 30, 2020
|
|
4X6A
 
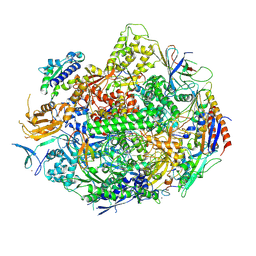 | | Crystal structure of yeast RNA polymerase II encountering oxidative Cyclopurine DNA lesions | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2014-12-07 | | Release date: | 2015-02-04 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3.96 Å) | | Cite: | Mechanism of RNA polymerase II bypass of oxidative cyclopurine DNA lesions.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8JX6
 
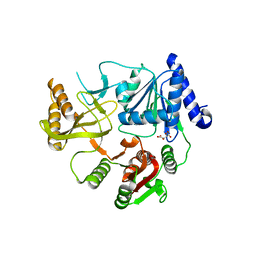 | | Deep-Sea Helicase 9 (DSH9) | | Descriptor: | Deep-Sea Helicase 9, L(+)-TARTARIC ACID, MAGNESIUM ION | | Authors: | Wang, L, Liu, Z, Guo, F. | | Deposit date: | 2023-06-30 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Deep-Sea Helicase 9 (DSH9)
To Be Published
|
|
4U7P
 
 | | Crystal structure of DNMT3A-DNMT3L complex | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Wang, L, Guo, X, Li, J, Xiao, J, Yin, X, He, S, Wang, J, Xu, Y. | | Deposit date: | 2014-07-31 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.821 Å) | | Cite: | Structural insight into autoinhibition and histone H3-induced activation of DNMT3A
Nature, 517, 2015
|
|
6L19
 
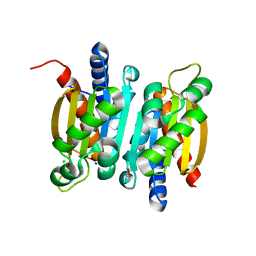 | | The crystal structure of competence or damage-inducible protein from Enterobacter asburiae | | Descriptor: | CHLORIDE ION, GLYCEROL, PncC family amidohydrolase, ... | | Authors: | Wang, L, Chen, Y, Liu, W, Lan, J, Shang, F, Xu, Y. | | Deposit date: | 2019-09-28 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The crystal structure of Competence or damage-inducible protein from Enterobacter asburiae
To Be Published
|
|
