8W8S
 
 | | Cryo-EM structure of the AA14-bound GPR101 complex | | Descriptor: | 1-(4-methylpyridin-2-yl)-3-[3-(trifluoromethyl)phenyl]thiourea, Probable G-protein coupled receptor 101 | | Authors: | Sun, J.P, Yu, X, Gao, N, Yang, F, Wang, J.Y, Yang, Z, Guan, Y, Wang, G.P. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
6YG7
 
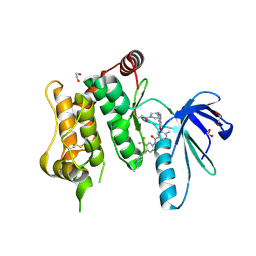 | | Crystal structure of MKK7 (MAP2K7) covalently bound with type-II inhibitor SB1-G-23 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity mitogen-activated protein kinase kinase 7, ~{N}-[4-[(4-ethylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl]-4-methyl-3-[2-[[(3~{R})-1-propanoylpyrrolidin-3-yl]amino]pyrimidin-4-yl]oxy-benzamide | | Authors: | Chaikuad, A, Tan, L, Wang, J, Liang, Y, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-27 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic Domain Plasticity of MKK7 Reveals Structural Mechanisms of Allosteric Activation and Diverse Targeting Opportunities.
Cell Chem Biol, 27, 2020
|
|
2BM2
 
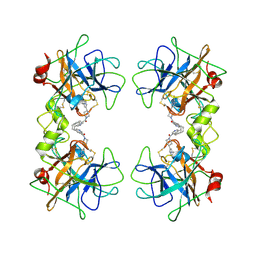 | | human beta-II tryptase in complex with 4-(3-Aminomethyl-phenyl)- piperidin-1-yl-(5-phenethyl- pyridin-3-yl)-methanone | | Descriptor: | 1-[3-(1-{[5-(2-PHENYLETHYL)PYRIDIN-3-YL]CARBONYL}PIPERIDIN-4-YL)PHENYL]METHANAMINE, HUMAN BETA2 TRYPTASE | | Authors: | Maignan, S, Guilloteau, J.-P, Dupuy, A, Levell, J, Astles, P, Eastwood, P, Cairns, J, Houille, O, Aldous, S, Merriman, G, Whiteley, B, Pribish, J, Czekaj, M, Liang, G, Davidson, J, Harrison, T, Morley, A, Watson, S, Fenton, G, Mccarthy, C, Romano, J, Mathew, R, Engers, D, Gardyan, M, Sides, K, Kwong, J, Tsay, J, Rebello, S, Shen, L, Wang, J, Luo, Y, Giardino, O, Lim, H.-K, Smith, K, Pauls, H. | | Deposit date: | 2005-03-09 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Based Design of 4-(3-Aminomethylphenyl) Piperidinyl-1-Amides: Novel, Potent, Selective, and Orally Bioavailable Inhibitors of Bii Tryptase
Bioorg.Med.Chem., 13, 2005
|
|
3ID4
 
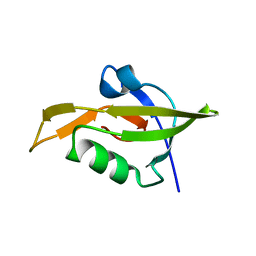 | | Crystal Structure of RseP PDZ2 domain fused GKASPV peptide | | Descriptor: | Regulator of sigma E protease | | Authors: | Li, X, Wang, B, Feng, L, Wang, J, Shi, Y. | | Deposit date: | 2009-07-20 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Cleavage of RseA by RseP requires a carboxyl-terminal hydrophobic amino acid following DegS cleavage
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3ID1
 
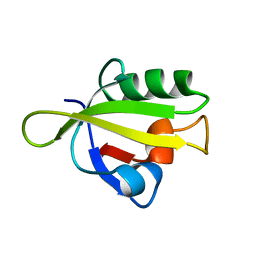 | | Crystal Structure of RseP PDZ1 domain | | Descriptor: | Regulator of sigma E protease | | Authors: | Li, X, Wang, B, Feng, L, Wang, J, Shi, Y. | | Deposit date: | 2009-07-20 | | Release date: | 2009-08-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Cleavage of RseA by RseP requires a carboxyl-terminal hydrophobic amino acid following DegS cleavage
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3MQ7
 
 | | Crystal Structure of Ectodomain Mutant of BST-2/Tetherin/CD317 | | Descriptor: | Bone marrow stromal antigen 2, CALCIUM ION | | Authors: | Xiong, Y, Yang, H, Wang, J, Meng, W. | | Deposit date: | 2010-04-27 | | Release date: | 2010-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural insight into the mechanisms of enveloped virus tethering by tetherin.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4YNZ
 
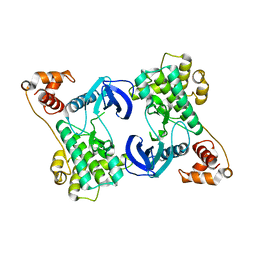 | | Structure of the N-terminal domain of SAD | | Descriptor: | Serine/threonine-protein kinase BRSK2 | | Authors: | Wu, J.X, Wang, J, Chen, L, Wang, Z.X, Wu, J.W. | | Deposit date: | 2015-03-11 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into the mechanism of synergistic autoinhibition of SAD kinases
Nat Commun, 6, 2015
|
|
4YOM
 
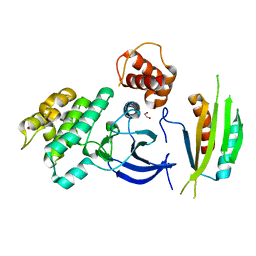 | | Structure of SAD kinase | | Descriptor: | 1,2-ETHANEDIOL, Serine/threonine-protein kinase BRSK2 | | Authors: | Wu, J.X, Wang, J, Chen, L, Wang, Z.X, Wu, J.W. | | Deposit date: | 2015-03-12 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural insight into the mechanism of synergistic autoinhibition of SAD kinases
Nat Commun, 6, 2015
|
|
6VZP
 
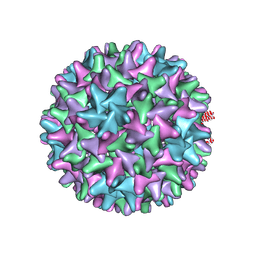 | | HBV wild type capsid | | Descriptor: | Capsid protein | | Authors: | Zhao, Z, Wang, J, Zlotnick, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-09-30 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The Integrity of the Intradimer Interface of the Hepatitis B Virus Capsid Protein Dimer Regulates Capsid Self-Assembly.
Acs Chem.Biol., 15, 2020
|
|
1D9K
 
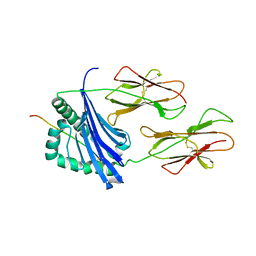 | | CRYSTAL STRUCTURE OF COMPLEX BETWEEN D10 TCR AND PMHC I-AK/CA | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CONALBUMIN PEPTIDE, ... | | Authors: | Reinherz, E.L, Tan, K, Tang, L, Kern, P, Liu, J.-H, Xiong, Y, Hussey, R.E, Smolyar, A, Hare, B, Zhang, R, Joachimiak, A, Chang, H.-C, Wagner, G, Wang, J.-H. | | Deposit date: | 1999-10-28 | | Release date: | 1999-12-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of a T cell receptor in complex with peptide and MHC class II.
Science, 286, 1999
|
|
6OMG
 
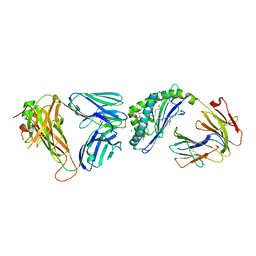 | | Structure of mouse CD1D- Glc-DAG (sn-1 C18:0, sn-2 C18:1c9)-iNKT TCR Ternary complex | | Descriptor: | (2R)-1-(alpha-D-glucopyranosyloxy)-3-(octadecanoyloxy)propan-2-yl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dirk, M.Z, Bitra, A, Wang, J. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of mouse CD1D- Glc-DAG (sn-1 C18:0, sn-2 C18:1c9)-iNKT TCR Ternary complex
To Be Published
|
|
6W0K
 
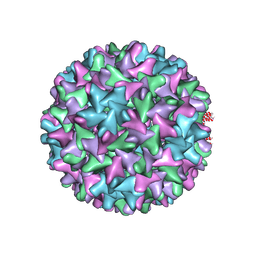 | | HBV D78S mutant capsid | | Descriptor: | Capsid protein | | Authors: | Zhao, Z, Wang, J, Zlotnick, A. | | Deposit date: | 2020-03-01 | | Release date: | 2020-09-30 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | The Integrity of the Intradimer Interface of the Hepatitis B Virus Capsid Protein Dimer Regulates Capsid Self-Assembly.
Acs Chem.Biol., 15, 2020
|
|
5MV1
 
 | | Crystal structure of the E protein of the Japanese encephalitis virulent virus | | Descriptor: | E protein | | Authors: | Liu, X, Zhao, X, Na, R, Li, L, Warkentin, E, Witt, J, Lu, X, Wei, Y, Peng, G, Li, Y, Wang, J. | | Deposit date: | 2017-01-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure differences of Japanese encephalitis virus SA14 and SA14-14-2 E proteins elucidate the virulence attenuation mechanism.
Protein Cell, 10, 2019
|
|
6DCG
 
 | | Discovery of MK-8353: An Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology | | Descriptor: | (3S)-3-(methylsulfanyl)-1-(2-{4-[4-(1-methyl-1H-1,2,4-triazol-3-yl)phenyl]-3,6-dihydropyridin-1(2H)-yl}-2-oxoethyl)-N-(3-{6-[(propan-2-yl)oxy]pyridin-3-yl}-1H-indazol-5-yl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Boga, S.B, Deng, Y, Zhu, L, Nan, Y, Cooper, A, Shipps Jr, G.W, Doll, R, Shih, N, Zhu, H, Sun, R, Wang, T, Paliwal, S, Tsui, H, Gao, X, Yao, X, Desai, J, Wang, J, Alhassan, A.B, Kelly, J, Patel, M, Muppalla, K, Gudipati, S, Zhang, L, Buevich, A, Hesk, D, Carr, D, Dayananth, P, Mei, H, Cox, K, Sherborne, B, Hruza, A.W, Xiao, L, Jin, W, Long, B, Liu, G, Taylor, S.A, Kirschmeier, P, Windsor, W.T, Bishop, R, Samatar, A.A. | | Deposit date: | 2018-05-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | MK-8353: Discovery of an Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology.
ACS Med Chem Lett, 9, 2018
|
|
5H64
 
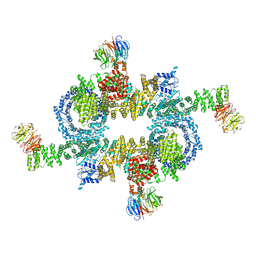 | | Cryo-EM structure of mTORC1 | | Descriptor: | Regulatory-associated protein of mTOR, Serine/threonine-protein kinase mTOR, Target of rapamycin complex subunit LST8 | | Authors: | Yang, H, Wang, J, Liu, M, Chen, X, Huang, M, Tan, D, Dong, M, Wong, C.C.L, Wang, J, Xu, Y, Wang, H. | | Deposit date: | 2016-11-10 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | 4.4 angstrom Resolution Cryo-EM structure of human mTOR Complex 1
Protein Cell, 7, 2016
|
|
3ID3
 
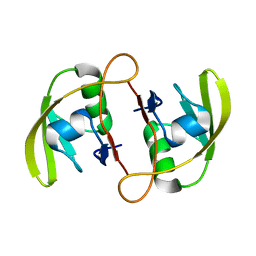 | | Crystal Structure of RseP PDZ2 I304A domain | | Descriptor: | Regulator of sigma E protease | | Authors: | Li, X, Wang, B, Feng, L, Wang, J, Shi, Y. | | Deposit date: | 2009-07-20 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Cleavage of RseA by RseP requires a carboxyl-terminal hydrophobic amino acid following DegS cleavage
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8W8Q
 
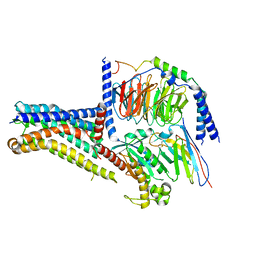 | | Cryo-EM structure of the GPR101-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Sun, J.P, Gao, N, Yu, X, Wang, G.P, Yang, F, Wang, J.Y, Yang, Z, Guan, Y. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
3CMZ
 
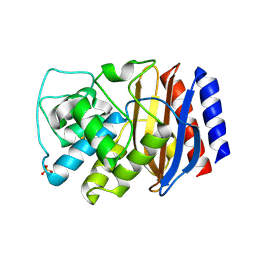 | | TEM-1 Class-A beta-lactamase L201P mutant apo structure | | Descriptor: | Beta-lactamase TEM, PHOSPHATE ION | | Authors: | Marciano, D.C, Wang, X, Wang, J, Chen, Y, Thomas, V.L, Shoichet, B.K, Palzkill, T. | | Deposit date: | 2008-03-24 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Genetic and structural characterization of an L201P global suppressor substitution in TEM-1 beta-lactamase
J.Mol.Biol., 384, 2008
|
|
3MQ9
 
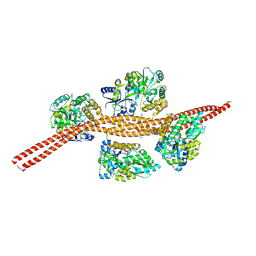 | |
3RGX
 
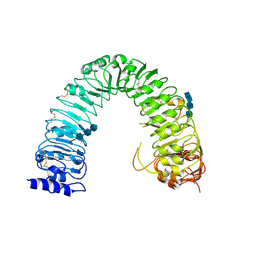 | | Structural insight into brassinosteroid perception by BRI1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chai, J, Han, Z, She, J, Wang, J, Cheng, W, Wang, J. | | Deposit date: | 2011-04-11 | | Release date: | 2011-06-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural insight into brassinosteroid perception by BRI1.
Nature, 474, 2011
|
|
1TAQ
 
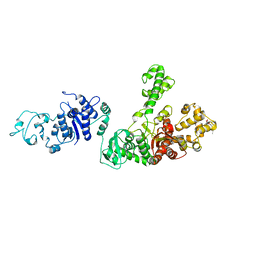 | | STRUCTURE OF TAQ DNA POLYMERASE | | Descriptor: | 2-O-octyl-beta-D-glucopyranose, TAQ DNA POLYMERASE, ZINC ION | | Authors: | Kim, Y, Eom, S.H, Wang, J, Lee, D.-S, Suh, S.W, Steitz, T.A. | | Deposit date: | 1996-06-04 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Thermus aquaticus DNA polymerase.
Nature, 376, 1995
|
|
5JPS
 
 | |
3K3N
 
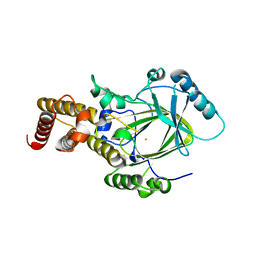 | | Crystal structure of the catalytic core domain of human PHF8 | | Descriptor: | FE (II) ION, PHD finger protein 8 | | Authors: | Yu, L, Wang, Y, Huang, S, Wang, J, Deng, Z, Wu, W, Gong, W, Chen, Z. | | Deposit date: | 2009-10-03 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into a novel histone demethylase PHF8
Cell Res., 20, 2010
|
|
2EFG
 
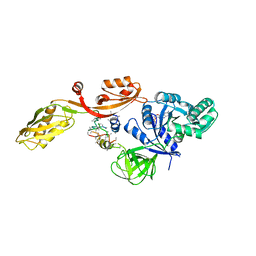 | | TRANSLATIONAL ELONGATION FACTOR G COMPLEXED WITH GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, PROTEIN (ELONGATION FACTOR G DOMAIN 3), PROTEIN (ELONGATION FACTOR G) | | Authors: | Czworkowski, J, Wang, J, Steitz, T.A, Moore, P.B. | | Deposit date: | 1998-09-23 | | Release date: | 1999-09-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of elongation factor G complexed with GDP, at 2.7 A resolution.
EMBO J., 13, 1994
|
|
5IAY
 
 | | NMR structure of UHRF1 Tandem Tudor Domains in a complex with Spacer peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Spacer | | Authors: | Fang, J, Cheng, J, Wang, J, Zhang, Q, Liu, M, Gong, R, Wang, P, Zhang, X, Feng, Y, Lan, W, Gong, Z, Tang, C, Wong, J, Yang, H, Cao, C, Xu, Y. | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition
Nat Commun, 7, 2016
|
|
