1U6B
 
 | | CRYSTAL STRUCTURE OF A SELF-SPLICING GROUP I INTRON WITH BOTH EXONS | | Descriptor: | 197-MER, 5'-R(*AP*AP*GP*CP*CP*AP*CP*AP*CP*AP*AP*AP*CP*CP*AP*GP*AP*CP*GP *GP*CP*C)-3', 5'-R(*CP*AP*(5MU))-3', ... | | Authors: | Adams, P.L, Stahley, M.R, Kosek, A.B, Wang, J, Strobel, S.A. | | Deposit date: | 2004-07-29 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of a Self-Splicing Group I Intron with Both Exons.
Nature, 430, 2004
|
|
4UIF
 
 | | Cryo-EM structure of Dengue virus serotype 2 in complex with antigen-binding fragments of human antibody 2D22 | | Descriptor: | ANTIGEN-BINDING FRAGMENT OF HUMAN ANTIBODY 2D22 - HEAVY CHAIN, ANTIGEN-BINDING FRAGMENT OF HUMAN ANTIBODY 2D22 - LIGHT CHAIN, DENGUE VIRUS SEROTYPE 2 STRAIN PVP94 07 - ENVELOPE PROTEIN, ... | | Authors: | Fibriansah, G, Ibarra, K.D, Ng, T.-S, Smith, S.A, Tan, J.L, Lim, X.-N, Ooi, J.S.G, Kostyuchenko, V.A, Wang, J, de Silva, A.M, Harris, E, Crowe Junior, J.E, Lok, S.-M. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers.
Science, 349, 2015
|
|
4UIH
 
 | | Cryo-EM structure of Dengue virus serotype 2 strain New Guinea-C complexed with human antibody 2D22 Fab at 37 degree C. The Fab molecules were added to the virus before 37 degree C incubation. | | Descriptor: | ANTIGEN-BINDING FRAGMENT OF HUMAN ANTIBODY 2D22 -HEAVY CHAIN, ANTIGEN-BINDING FRAGMENT OF HUMAN ANTIBODY 2D22 -LIGHT CHAIN, DENGUE VIRUS SEROTYPE 2 STRAIN NEW GUINEA-C E PROTEIN ECTODOMAIN | | Authors: | Fibriansah, G, Ibarra, K.D, Ng, T.-S, Smith, S.A, Tan, J.L, Lim, X.N, Ooi, J.S.G, Kostyuchenko, V.A, Wang, J, de Silva, A.M, Harris, E, Crowe, J.E, Lok, S.-M. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers.
Science, 349, 2015
|
|
8D8O
 
 | | Cryo-EM structure of substrate unbound PAPP-A | | Descriptor: | Pappalysin-1, ZINC ION | | Authors: | Judge, R.A, Jain, R, Hao, Q, Ouch, C, Sridar, J, Smith, C.L, Wang, J.C.K, Eaton, D. | | Deposit date: | 2022-06-08 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure of the PAPP-ABP5 complex reveals mechanism of substrate recognition
Nat Commun, 13, 2022
|
|
2Y1R
 
 | | Structure of MecA121 & ClpC N-domain complex | | Descriptor: | ADAPTER PROTEIN MECA 1, NEGATIVE REGULATOR OF GENETIC COMPETENCE CLPC/MECB, S,R MESO-TARTARIC ACID | | Authors: | Wang, F, Mei, Z.Q, Wang, J.W, Shi, Y.G. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Structure and Mechanism of the Hexameric Meca-Clpc Molecular Machine.
Nature, 471, 2011
|
|
5JCD
 
 | | Crystal structure of OsCEBiP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor-binding protein | | Authors: | Chai, J.J, Liu, S.M, Wang, J.Z. | | Deposit date: | 2016-04-15 | | Release date: | 2017-02-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Mechanism for Fungal Cell Wall Recognition by Rice Chitin Receptor OsCEBiP
Structure, 24, 2016
|
|
7TUK
 
 | |
7TUL
 
 | |
6T7K
 
 | | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline--tRNA ligase) from Plasmodium falciparum in complex with NCP-26 and L-Proline | | Descriptor: | 1,2-ETHANEDIOL, PROLINE, Proline--tRNA ligase, ... | | Authors: | Johansson, C, Wang, J, Tye, M, Payne, N.C, Mazitschek, R, Thompson, A, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2019-10-22 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline--tRNA ligase) from Plasmodium falciparum in complex with NCP-26 and L-Proline
To Be Published
|
|
1L0Q
 
 | | Tandem YVTN beta-propeller and PKD domains from an archaeal surface layer protein | | Descriptor: | Surface layer protein | | Authors: | Jing, H, Takagi, J, Liu, J.-H, Lindgren, S, Zhang, R.-G, Joachimiak, A, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-02-12 | | Release date: | 2002-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Archaeal Surface Layer Proteins Contain beta Propeller, PKD, and beta Helix Domains and Are Related to Metazoan Cell Surface Proteins.
Structure, 10, 2002
|
|
1DKC
 
 | | SOLUTION STRUCTURE OF PAFP-S, AN ANTIFUNGAL PEPTIDE FROM THE SEEDS OF PHYTOLACCA AMERICANA | | Descriptor: | ANTIFUNGAL PEPTIDE | | Authors: | Wang, D.C, Gao, G.H, Shao, F, Dai, J.X, Wang, J.F. | | Deposit date: | 1999-12-07 | | Release date: | 2000-12-13 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PAFP-S: a new knottin-type antifungal peptide from the seeds of Phytolacca americana
Biochemistry, 40, 2001
|
|
6WFS
 
 | | Cryo-EM Structure of Hepatitis B virus T=4 capsid in complex with the antiviral molecule DBT1 | | Descriptor: | 11-oxo-N-[2-(4-sulfamoylphenyl)ethyl]-10,11-dihydrodibenzo[b,f][1,4]thiazepine-8-carboxamide, Capsid protein | | Authors: | Schlicksup, C, Laughlin, P, Dunkelbarger, S, Wang, J.C, Zlotnick, A. | | Deposit date: | 2020-04-03 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Local Stabilization of Subunit-Subunit Contacts Causes Global Destabilization of Hepatitis B Virus Capsids.
Acs Chem.Biol., 15, 2020
|
|
6WGJ
 
 | | Fab portion of dupilumab with Crystal Kappa design and no interchain disulfide | | Descriptor: | Dupilumab Fab heavy chain, Dupilumab Fab light chain | | Authors: | Druzina, Z, Atwell, S, Pustilnik, A, Antonysamy, S, Ho, C, Lieu, R, Hendle, J, Benach, J, Wang, J. | | Deposit date: | 2020-04-05 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rapid and robust antibody Fab fragment crystallization utilizing edge-to-edge beta-sheet packing.
Plos One, 15, 2020
|
|
5C4W
 
 | | Crystal structure of coxsackievirus A16 | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SODIUM ION, ... | | Authors: | Ren, J, Wang, X, Zhu, L, Hu, Z, Gao, Q, Yang, P, Li, X, Wang, J, Shen, X, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2015-06-18 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of Coxsackievirus A16 Capsids with Native Antigenicity: Implications for Particle Expansion, Receptor Binding, and Immunogenicity.
J.Virol., 89, 2015
|
|
5C9A
 
 | | Crystal structure of empty coxsackievirus A16 particle | | Descriptor: | CHLORIDE ION, POTASSIUM ION, SPHINGOSINE, ... | | Authors: | Ren, J, Wang, X, Zhu, L, Hu, Z, Gao, Q, Yang, P, Li, X, Wang, J, Shen, X, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2015-06-26 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of Coxsackievirus A16 Capsids with Native Antigenicity: Implications for Particle Expansion, Receptor Binding, and Immunogenicity.
J.Virol., 89, 2015
|
|
6M1I
 
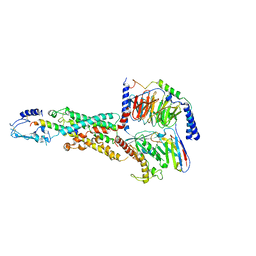 | | CryoEM structure of human PAC1 receptor in complex with PACAP38 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Song, X, Wang, J, Zhang, D, Wang, H.W, Ma, Y. | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism.
Cell Res., 30, 2020
|
|
5MGV
 
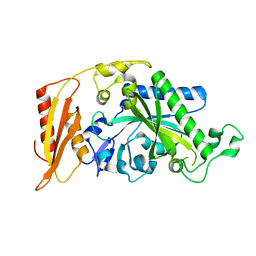 | | Kinetic and Structural Changes in HsmtPheRS, Induced by Pathogenic Mutations in Human FARS2 | | Descriptor: | Phenylalanine--tRNA ligase, mitochondrial | | Authors: | Kartvelishvili, E, Tworowski, D, Vernon, H, Chrzanowska-Lightowlers, Z, Moor, N, Wang, J, Wong, L.-J, Safro, M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-05-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Kinetic and structural changes in HsmtPheRS, induced by pathogenic mutations in human FARS2.
Protein Sci., 26, 2017
|
|
4RHR
 
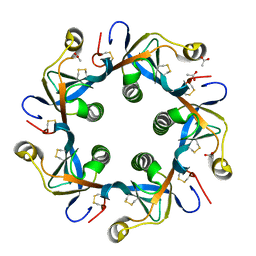 | | Crystal structure of PltB | | Descriptor: | ACETATE ION, Putative pertussis-like toxin subunit | | Authors: | Gao, X, Wang, J, Galan, J. | | Deposit date: | 2014-10-02 | | Release date: | 2014-10-29 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Host adaptation of a bacterial toxin from the human pathogen salmonella typhi.
Cell(Cambridge,Mass.), 159, 2014
|
|
4RHS
 
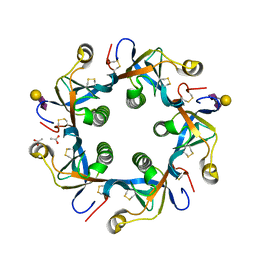 | | Crystal structure of GD2 bound PltB | | Descriptor: | ACETATE ION, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, Putative pertussis-like toxin subunit | | Authors: | Gao, X, Wang, J, Galan, J. | | Deposit date: | 2014-10-02 | | Release date: | 2014-10-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9211 Å) | | Cite: | Host adaptation of a bacterial toxin from the human pathogen salmonella typhi.
Cell(Cambridge,Mass.), 159, 2014
|
|
6XA4
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with UAW241 | | Descriptor: | 3C-like proteinase, GLYCEROL, inhibitor UAW241 | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
1SZT
 
 | |
5MGU
 
 | | Kinetic and Structural Changes in HsmtPheRS, Induced by Pathogenic Mutations in Human FARS2 | | Descriptor: | PHENYLALANINE, Phenylalanine--tRNA ligase, mitochondrial | | Authors: | Kartvelishvili, E, Tworowski, D, Vernon, H, Chrzanowska-Lightowlers, Z, Moor, N, Wang, J, Wong, L.-J, Safro, M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-05-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Kinetic and structural changes in HsmtPheRS, induced by pathogenic mutations in human FARS2.
Protein Sci., 26, 2017
|
|
5MGH
 
 | | Crystal structure of pathogenic mutants of human mitochodnrial PheRS | | Descriptor: | PHENYLALANINE, Phenylalanine--tRNA ligase, mitochondrial | | Authors: | Kartvelishvili, E, Tworowski, D, Vernon, H, Chrzanowska-Lightowlers, Z, Moor, N, Wang, J, Wong, L.-J, Safro, M. | | Deposit date: | 2016-11-21 | | Release date: | 2017-05-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Kinetic and structural changes in HsmtPheRS, induced by pathogenic mutations in human FARS2.
Protein Sci., 26, 2017
|
|
6XF4
 
 | | Crystal structure of STING REF variant in complex with E7766 | | Descriptor: | (1R,3R,15E,28R,29R,30R,31R,34R,36R,39S,41R)-29,41-difluoro-34,39-disulfanyl-2,33,35,38,40,42-hexaoxa-4,6,9,11,13,18,20,22,25,27-decaaza-34,39-diphosphaoctacyclo[28.6.4.1~3,36~.1~28,31~.0~4,8~.0~7,12~.0~19,24~.0~23,27~]dotetraconta-5,7,9,11,15,19,21,23,25-nonaene 34,39-dioxide (non-preferred name), 1,2-ETHANEDIOL, Stimulator of interferon genes protein | | Authors: | Chen, Y, Wang, J.Y, Kim, D.-S. | | Deposit date: | 2020-06-15 | | Release date: | 2021-02-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | E7766, a Macrocycle-Bridged Stimulator of Interferon Genes (STING) Agonist with Potent Pan-Genotypic Activity.
Chemmedchem, 16, 2021
|
|
6XF3
 
 | | Crystal structure of STING in complex with E7766 | | Descriptor: | (1R,3R,15E,28R,29R,30R,31R,34R,36R,39S,41R)-29,41-difluoro-34,39-disulfanyl-2,33,35,38,40,42-hexaoxa-4,6,9,11,13,18,20,22,25,27-decaaza-34,39-diphosphaoctacyclo[28.6.4.1~3,36~.1~28,31~.0~4,8~.0~7,12~.0~19,24~.0~23,27~]dotetraconta-5,7,9,11,15,19,21,23,25-nonaene 34,39-dioxide (non-preferred name), Stimulator of interferon genes protein | | Authors: | Chen, Y, Wang, J.Y, Kim, D.-S. | | Deposit date: | 2020-06-15 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | E7766, a Macrocycle-Bridged Stimulator of Interferon Genes (STING) Agonist with Potent Pan-Genotypic Activity.
Chemmedchem, 16, 2021
|
|
