7DAC
 
 | | Human RIPK3 amyloid fibril revealed by solid-state NMR | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Wu, X.L, Zhang, J, Dong, X.Q, Liu, J, Li, B, Hu, H, Wang, J, Wang, H.Y, Lu, J.X. | | Deposit date: | 2020-10-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | The structure of a minimum amyloid fibril core formed by necroptosis-mediating RHIM of human RIPK3.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8G9D
 
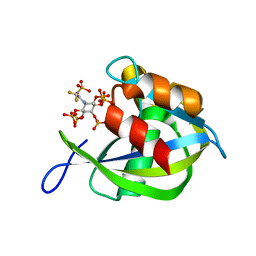 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with 5- phosphonodifluoroacetamide inositol pentakisphosphate (5-PCF2Am-InsP5), an analogue of 5-InsP7 | | Descriptor: | (1,1-difluoro-2-oxo-2-{[(1s,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl]amino}ethyl)phosphonic acid, Diphosphoinositol polyphosphate phosphohydrolase 1 | | Authors: | Zong, G, Wang, H, Shears, S. | | Deposit date: | 2023-02-21 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fluorination Influences the Bioisostery of Myo-Inositol Pyrophosphate Analogs.
Chemistry, 29, 2023
|
|
4YZF
 
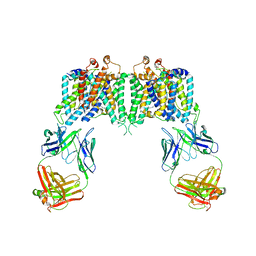 | | Crystal structure of the anion exchanger domain of human erythrocyte Band 3 | | Descriptor: | 2,2'-ethane-1,2-diylbis{5-[(sulfanylmethyl)amino]benzenesulfonic acid}, Band 3 anion transport protein, FAB fragment of Immunoglobulin (IgG) molecule | | Authors: | Alguel, Y, Arakawa, T, Yugiri, T.K, Iwanari, H, Hatae, H, Iwata, M, Abe, Y, Hino, T, Suno, C.I, Kuma, H, Kang, D, Murata, T, Hamakubo, T, Cameron, A.D, Kobayashi, T, Hamasaki, N, Iwata, S. | | Deposit date: | 2015-03-25 | | Release date: | 2015-11-04 | | Last modified: | 2015-11-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the anion exchanger domain of human erythrocyte band 3.
Science, 350, 2015
|
|
5A16
 
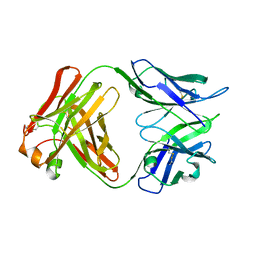 | | Crystal structure of Fab4201 raised against Human Erythrocyte Anion Exchanger 1 | | Descriptor: | FAB4201 HEAVY CHAIN | | Authors: | Arakawa, T, Kobayashi-Yugiri, T, Alguel, Y, Weyand, S, Iwanari, H, Hatae, H, Iwata, M, Abe, Y, Hino, T, Ikeda-Suno, C, Kuma, H, Kang, D, Murata, T, Hamakubo, T, Cameron, A, Kobayashi, T, Hamasaki, N, Iwata, S. | | Deposit date: | 2015-04-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Anion Exchanger Domain of Human Erythrocyte Band 3
Science, 350, 2015
|
|
4FJ4
 
 | | Crystal structure of the protein Q9HRE7 complexed with mercury from Halobacterium salinarium at the resolution 2.1A, Northeast Structural Genomics Consortium target HsR50 | | Descriptor: | ETHYL MERCURY ION, SODIUM ION, Uncharacterized protein | | Authors: | Kuzin, A, Chen, Y, Vorobiev, S.M, Seetharaman, J, Janjua, J, Xiao, R, Cunningham, K, Maglaqui, M, Owens, L.A, Wang, H, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-11 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Northeast Structural Genomics Consortium Target HsR50
To be Published
|
|
5ZOO
 
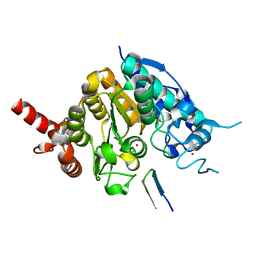 | | Crystal structure of histone deacetylase 4 (HDAC4) in complex with a SMRT corepressor SP1 fragment | | Descriptor: | Histone deacetylase 4, POTASSIUM ION, SMRT corepressor SP1 fragment, ... | | Authors: | Park, S.Y, Hwang, H.J, Kim, J.S. | | Deposit date: | 2018-04-13 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of the specific interaction of SMRT corepressor with histone deacetylase 4.
Nucleic Acids Res., 46, 2018
|
|
2Z8M
 
 | | Structural basis for the catalytic mechanism of phosphothreonine lyase | | Descriptor: | 27.5 kDa virulence protein | | Authors: | Chen, L, Wang, H, Gu, L, Huang, N, Zhou, J.M, Chai, J. | | Deposit date: | 2007-09-07 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the catalytic mechanism of phosphothreonine lyase.
Nat.Struct.Mol.Biol., 15, 2008
|
|
1VLZ
 
 | |
3TYH
 
 | | Crystal structure of oxo-cupper clusters binding to ferric binding protein from Neisseria gonorrhoeae | | Descriptor: | COPPER (II) ION, FbpA protein | | Authors: | Chen, W.J, Wang, H.F, Zhou, C.J, Ye, D.R, Huang, J, Tan, X.S, Zhong, W.Q. | | Deposit date: | 2011-09-26 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of oxo-cupper clusters binding to ferric binding protein from Neisseria gonorrhoeae
To be Published
|
|
7EVP
 
 | | Cryo-EM structure of the Gp168-beta-clamp complex | | Descriptor: | Beta sliding clamp, Sliding clamp inhibitor | | Authors: | Liu, B, Li, S, Liu, Y, Chen, H, Hu, Z, Wang, Z, Gou, L, Zhang, L, Ma, B, Wang, H, Matthews, S, Wang, Y, Zhang, K. | | Deposit date: | 2021-05-21 | | Release date: | 2022-02-16 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Bacteriophage Twort protein Gp168 is a beta-clamp inhibitor by occupying the DNA sliding channel.
Nucleic Acids Res., 49, 2021
|
|
3J0A
 
 | | Homology model of human Toll-like receptor 5 fitted into an electron microscopy single particle reconstruction | | Descriptor: | Toll-like receptor 5, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Modis, Y, Zhou, K, Kanai, R, Lee, P, Wang, H.W. | | Deposit date: | 2011-06-02 | | Release date: | 2011-12-28 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (26 Å) | | Cite: | Toll-like receptor 5 forms asymmetric dimers in the absence of flagellin.
J.Struct.Biol., 177, 2012
|
|
7FH6
 
 | | Friedel-Crafts alkylation enzyme CylK | | Descriptor: | CALCIUM ION, CHLORIDE ION, CylK, ... | | Authors: | Liu, L, Wang, H.Q, Xiang, Z. | | Deposit date: | 2021-07-29 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for the Friedel-Crafts Alkylation in Cylindrocyclophane Biosynthesis
ACS Catal., 12, 2022
|
|
4Z5T
 
 | | The nucleosome containing human H3.5 | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Urahama, T, Harada, A, Maehara, K, Horikoshi, N, Sato, K, Sato, Y, Shiraishi, K, Sugino, N, Osakabe, A, Tachiwana, H, Kagawa, W, Kimura, H, Ohkawa, Y, Kurumizaka, H. | | Deposit date: | 2015-04-03 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Histone H3.5 forms an unstable nucleosome and accumulates around transcription start sites in human testis.
Epigenetics Chromatin, 9, 2016
|
|
3E23
 
 | | Crystal structure of the RPA2492 protein in complex with SAM from Rhodopseudomonas palustris, Northeast Structural Genomics Consortium Target RpR299 | | Descriptor: | S-ADENOSYLMETHIONINE, SULFATE ION, uncharacterized protein RPA2492 | | Authors: | Forouhar, F, Chen, Y, Seetharaman, J, Mao, L, Xiao, R, Foote, E.L, Ciccosanti, C, Wang, H, Tong, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-08-05 | | Release date: | 2008-09-30 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the RPA2492 protein in complex with SAM from Rhodopseudomonas palustris, Northeast Structural Genomics Consortium Target RpR299
To be Published
|
|
2Z8P
 
 | | Structural basis for the catalytic mechanism of phosphothreonine lyase | | Descriptor: | (GLY)(GLU)(ALA)(TPO)(VAL)(PTR)(ALA), 27.5 kDa virulence protein | | Authors: | Chen, L, Wang, H, Gu, L, Huang, N, Zhou, J.M, Chai, J. | | Deposit date: | 2007-09-07 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the catalytic mechanism of phosphothreonine lyase.
Nat.Struct.Mol.Biol., 15, 2008
|
|
8T7H
 
 | | Quis-bound intermediate mGlu5 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Metabotropic glutamate receptor 5, Nanobody 43 | | Authors: | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | Deposit date: | 2023-06-20 | | Release date: | 2023-10-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Stepwise activation of a metabotropic glutamate receptor.
Nature, 629, 2024
|
|
8TAO
 
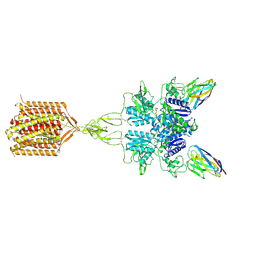 | | Quis and CDPPB bound active mGlu5 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 3-cyano-N-(1,3-diphenyl-1H-pyrazol-5-yl)benzamide, Metabotropic glutamate receptor 5, ... | | Authors: | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | Deposit date: | 2023-06-27 | | Release date: | 2023-10-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Stepwise activation of a metabotropic glutamate receptor.
Nature, 629, 2024
|
|
1BC9
 
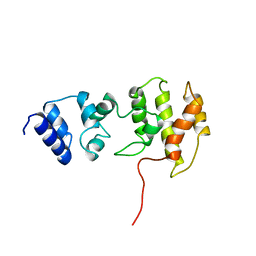 | | CYTOHESIN-1/B2-1 SEC7 DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOHESIN-1 | | Authors: | Betz, S.F, Schnuchel, A, Wang, H, Olejniczak, E.T, Meadows, R.P, Fesik, S.W. | | Deposit date: | 1998-05-06 | | Release date: | 1999-05-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cytohesin-1 (B2-1) Sec7 domain and its interaction with the GTPase ADP ribosylation factor 1.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
5B24
 
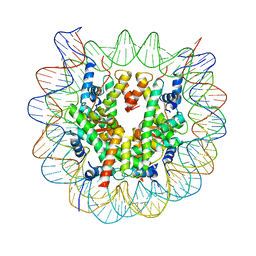 | | The crystal structure of the nucleosome containing cyclobutane pyrimidine dimer | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Horikoshi, N, Tachiwana, H, Kagawa, W, Osakabe, A, Kurumizaka, H. | | Deposit date: | 2015-12-31 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of the nucleosome containing ultraviolet light-induced cyclobutane pyrimidine dimer
Biochem.Biophys.Res.Commun., 471, 2016
|
|
7DIE
 
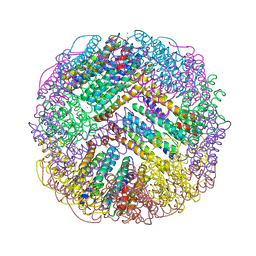 | | Crystal structure of M. penetrans Ferritin | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Wang, w.m, Zhang, y, Wang, h.f. | | Deposit date: | 2020-11-19 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ferritin with Atypical Ferroxidase Centers Takes B-Channels as the Pathway for Fe 2+ Uptake from Mycoplasma .
Inorg.Chem., 60, 2021
|
|
2LG2
 
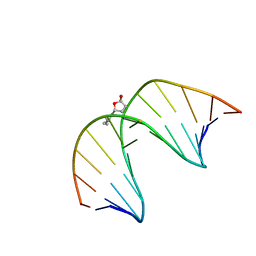 | | Structure of the duplex containing HNE derived (6S,8R,11S) N2-dG cyclic hemiacetal when placed opposite dT | | Descriptor: | (2R,5S)-5-pentyltetrahydrofuran-2-ol, DNA (5'-D(*GP*CP*TP*AP*GP*CP*GP*AP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*TP*TP*GP*CP*TP*AP*GP*C)-3') | | Authors: | Huang, H, Wang, H, Kozekova, A, Lloyd, R.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2011-07-19 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ring-chain tautomerization of trans-4-hydroxynonenal derived (6S,8R,11S) gamma-hydroxy-1,N2-propano-deoxyguanosine adduct when placed opposite 2'-deoxythymidine in duplex
To be Published
|
|
3UQH
 
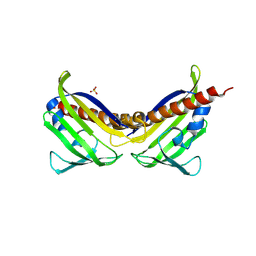 | | Crystal structure of aba receptor pyl10 (apo) | | Descriptor: | Abscisic acid receptor PYL10, SULFATE ION | | Authors: | Sun, D.M, Wang, H.P, Wu, M.H, Zang, J.Y, Tian, C.L. | | Deposit date: | 2011-11-20 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Aba Receptor Pyl10
To be Published
|
|
6L49
 
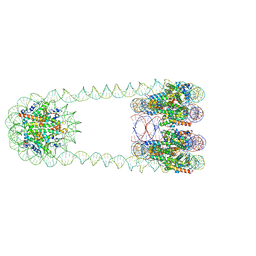 | | H3-CA-H3 tri-nucleosome with the 22 base-pair linker DNA | | Descriptor: | DNA (485-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Takizawa, Y, Ho, C.-H, Tachiwana, H, Matsunami, H, Ohi, M, Wolf, M, Kurumizaka, H. | | Deposit date: | 2019-10-16 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (18.9 Å) | | Cite: | Cryo-EM Structures of Centromeric Tri-nucleosomes Containing a Central CENP-A Nucleosome.
Structure, 28, 2020
|
|
2QLK
 
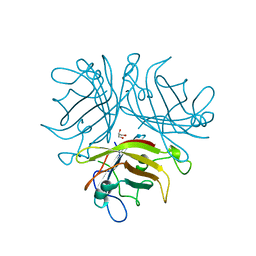 | | Adenovirus AD35 fibre head | | Descriptor: | Fiber, GLYCEROL | | Authors: | Liaw, Y.-C, Amiraslanov, I, Wang, H, Lieber, A. | | Deposit date: | 2007-07-13 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Identification of CD46 binding sites within the adenovirus serotype 35 fiber knob
J.Virol., 81, 2007
|
|
4RHP
 
 | | Crystal structure of human COQ9 in complex with a phospholipid, Northeast Structural Genomics Consortium Target HR5043 | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Ubiquinone biosynthesis protein COQ9, mitochondrial | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Wang, H, Lee, D, Kogan, S, Maglaqui, M, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2014-10-02 | | Release date: | 2014-10-22 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
