6IMH
 
 | | Solution Structure of Bicyclic Peptide pb-18 | | Descriptor: | (ACE)-GLY-CYS-PRO-CYS-GLU-PRO-SER-TYR-LEU-CYS-PRO-TRP-LEU-PRO-GLY-CYS-(NH2) | | Authors: | Yao, H, Lin, P, Zha, J, Zha, M, Zhao, Y, Wu, C. | | Deposit date: | 2018-10-22 | | Release date: | 2019-08-28 | | Method: | SOLUTION NMR | | Cite: | Ordered and Isomerically Stable Bicyclic Peptide Scaffolds Constrained through Cystine Bridges and Proline Turns.
Chembiochem, 20, 2019
|
|
1PJW
 
 | | Solution Structure of the Domain III of the Japan Encephalitis Virus Envelope Protein | | Descriptor: | envelope protein | | Authors: | Wu, K.P, Wu, C.W, Tsao, Y.P, Lou, Y.C, Lin, C.W. | | Deposit date: | 2003-06-04 | | Release date: | 2003-11-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of a Flavivirus Recognized by Its Neutralizing Antibody: SOLUTION STRUCTURE OF THE DOMAIN III OF THE JAPANESE ENCEPHALITIS VIRUS ENVELOPE PROTEIN.
J.Biol.Chem., 278, 2003
|
|
4YJ5
 
 | | Crystal structure of PKM2 mutant | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, PYRUVIC ACID, ... | | Authors: | Liu, J.-S, Wu, C.-W, Wang, W.-C. | | Deposit date: | 2015-03-03 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Mutations in the PKM2 exon-10 region are associated with reduced allostery and increased nuclear translocation
Commun Biol, 2, 2019
|
|
6YPZ
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
6YQ6
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
7RSJ
 
 | | Structure of the VPS34 kinase domain with compound 14 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-{4-[(7R,8R)-4-oxo-7-(propan-2-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazin-2-yl]pyridin-2-yl}cyclopropanecarboxamide, ... | | Authors: | Hu, D.X, Patel, S, Chen, H, Wang, S, Staben, S, Dimitrova, Y.N, Wallweber, H.A, Lee, J.Y, Chan, G.K.Y, Sneeringer, C.J, Prangley, M.S, Moffat, J.G, Wu, C, Schutt, L.K, Salphati, L, Pang, J, McNamara, E, Huang, H, Chen, Y, Wang, Y, Zhao, W, Lim, J, Murthy, A, Siu, M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | Structure-Based Design of Potent, Selective, and Orally Bioavailable VPS34 Kinase Inhibitors.
J.Med.Chem., 65, 2022
|
|
6YQ3
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | (3~{R})-8-methoxy-3-methyl-3,6-bis(oxidanyl)-2,4-dihydrobenzo[a]anthracene-1,7,12-trione, 1,2-ETHANEDIOL, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
7RSP
 
 | | Structure of the VPS34 kinase domain with compound 14 | | Descriptor: | (7R,8R)-2-[(3R)-3-methylmorpholin-4-yl]-7-(propan-2-yl)-6,7-dihydropyrazolo[1,5-a]pyrazin-4(5H)-one, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3 | | Authors: | Hu, D.X, Patel, S, Chen, H, Wang, S, Staben, S, Dimitrova, Y.N, Wallweber, H.A, Lee, J.Y, Chan, G.K.Y, Sneeringer, C.J, Prangley, M.S, Moffat, J.G, Wu, C, Schutt, L.K, Salphati, L, Pang, J, McNamara, E, Huang, H, Chen, Y, Wang, Y, Zhao, W, Lim, J, Murthy, A, Siu, M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure-Based Design of Potent, Selective, and Orally Bioavailable VPS34 Kinase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7RSV
 
 | | Structure of the VPS34 kinase domain with compound 5 | | Descriptor: | (5aS,8aR,9S)-2-[(3R)-3-methylmorpholin-4-yl]-5,5a,6,7,8,8a-hexahydro-4H-cyclopenta[e]pyrazolo[1,5-a]pyrazin-4-one, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3, ... | | Authors: | Hu, D.X, Patel, S, Chen, H, Wang, S, Staben, S, Dimitrova, Y.N, Wallweber, H.A, Lee, J.Y, Chan, G.K.Y, Sneeringer, C.J, Prangley, M.S, Moffat, J.G, Wu, C, Schutt, L.K, Salphati, L, Pang, J, McNamara, E, Huang, H, Chen, Y, Wang, Y, Zhao, W, Lim, J, Murthy, A, Siu, M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-Based Design of Potent, Selective, and Orally Bioavailable VPS34 Kinase Inhibitors.
J.Med.Chem., 65, 2022
|
|
1OQC
 
 | |
8SZG
 
 | | Cryo-EM structure of cinacalcet-bound human calcium-sensing receptor CaSR-Gq complex in lipid nanodiscs | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | He, F, Wu, C, Gao, Y, Skiniotis, G. | | Deposit date: | 2023-05-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Allosteric modulation and G-protein selectivity of the Ca 2+ -sensing receptor.
Nature, 626, 2024
|
|
7R81
 
 | | Structure of the translating Neurospora crassa ribosome arrested by cycloheximide | | Descriptor: | 18S rRNA, 26S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Shen, L, Su, Z, Yang, K, Wu, C, Becker, T, Bell-Pedersen, D, Zhang, J, Sachs, M.S. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of the translating Neurospora ribosome arrested by cycloheximide
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6XAF
 
 | | 1.9A crystal structure of the GTPase domain of Parkinson's disease-associated protein LRRK2 carrying R1398H | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2, MAGNESIUM ION | | Authors: | Hoang, Q.Q, Liao, J, Huang, X, Park, Y, Wu, C.X. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | Structural basis for conformational plasticity in the GTPase domain of the Parkinson's disease-associated protein LRRK2
To be Published
|
|
8SZH
 
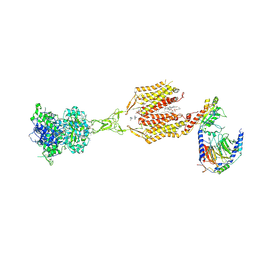 | | Cryo-EM structure of cinacalcet-bound human calcium-sensing receptor CaSR-Gi complex in lipid nanodiscs | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | He, F, Wu, C, Gao, Y, Skiniotis, G. | | Deposit date: | 2023-05-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Allosteric modulation and G-protein selectivity of the Ca 2+ -sensing receptor.
Nature, 626, 2024
|
|
1FSY
 
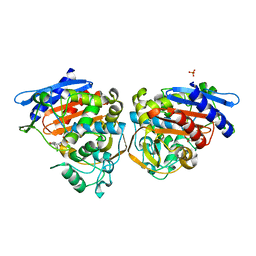 | | AMPC BETA-LACTAMASE FROM E. COLI COMPLEXED WITH INHIBITOR CLOXACILLINBORONIC ACID | | Descriptor: | CEPHALOSPORINASE, N-[5-METHYL-3-O-TOLYL-ISOXAZOLE-4-CARBOXYLIC ACID AMIDE] BORONIC ACID, PHOSPHATE ION | | Authors: | Caselli, E, Powers, R.A, Blasczcak, L.C, Wu, C.Y, Prati, F, Shoichet, B.K. | | Deposit date: | 2000-09-11 | | Release date: | 2001-03-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Energetic, structural, and antimicrobial analyses of beta-lactam side chain recognition by beta-lactamases.
Chem.Biol., 8, 2001
|
|
1FSW
 
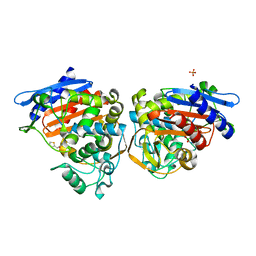 | | AMPC BETA-LACTAMASE FROM E. COLI COMPLEXED WITH INHIBITOR CEPHALOTHINBORONIC ACID | | Descriptor: | CEPHALOSPORINASE, N-2-THIOPHEN-2-YL-ACETAMIDE BORONIC ACID, PHOSPHATE ION | | Authors: | Caselli, E, Powers, R.A, Blaszczak, L.C, Wu, C.Y, Prati, F, Shoichet, B.K. | | Deposit date: | 2000-09-11 | | Release date: | 2001-03-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Energetic, structural, and antimicrobial analyses of beta-lactam side chain recognition by beta-lactamases.
Chem.Biol., 8, 2001
|
|
8SZI
 
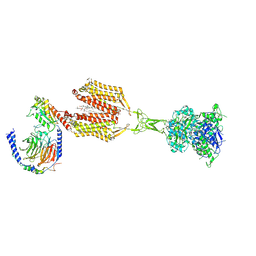 | | Cryo-EM structure of PAM-free human calcium-sensing receptor CaSR-Gi complex in lipid nanodiscs | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | He, F, Wu, C, Gao, Y, Skiniotis, G. | | Deposit date: | 2023-05-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Allosteric modulation and G-protein selectivity of the Ca 2+ -sensing receptor.
Nature, 626, 2024
|
|
8WSS
 
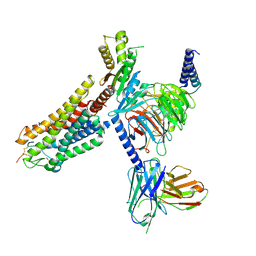 | | Cryo-EM structure of Melanin-Concentrating Hormone Receptor 1 with MCH | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhao, L, He, Q, Yuan, Q, Gu, Y, Shan, H, Hu, W, Wu, K, Xu, H.E, Zhang, Y, Wu, C. | | Deposit date: | 2023-10-17 | | Release date: | 2024-06-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Mechanisms of ligand recognition and activation of melanin-concentrating hormone receptors.
Cell Discov, 10, 2024
|
|
8WST
 
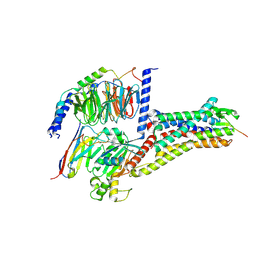 | | Cryo-EM structure of Melanin-Concentrating Hormone Receptor 2 with MCH | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha-1, ... | | Authors: | Zhao, L, He, Q, Yuan, Q, Gu, Y, Shan, H, Hu, W, Wu, K, Xu, H.E, Zhang, Y, Wu, C. | | Deposit date: | 2023-10-17 | | Release date: | 2024-06-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Mechanisms of ligand recognition and activation of melanin-concentrating hormone receptors.
Cell Discov, 10, 2024
|
|
8SZF
 
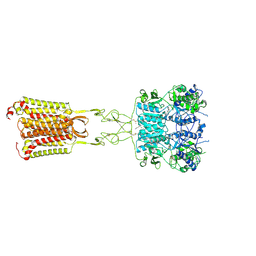 | | Cryo-EM structure of cinacalcet-bound active-state human calcium-sensing receptor CaSR in lipid nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | He, F, Wu, C, Gao, Y, Skiniotis, G. | | Deposit date: | 2023-05-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Allosteric modulation and G-protein selectivity of the Ca 2+ -sensing receptor.
Nature, 626, 2024
|
|
2PO7
 
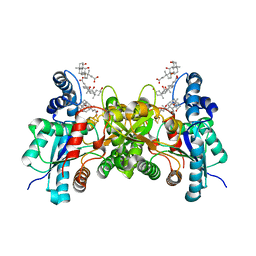 | | Crystal structure of human ferrochelatase mutant with His 341 replaced by Cys | | Descriptor: | CHOLIC ACID, FE2/S2 (INORGANIC) CLUSTER, Ferrochelatase, ... | | Authors: | Dailey, H.A, Wu, C.-K, Horanyi, P, Medlock, A.E, Najahi-Missaoui, W, Burden, A, Dailey, T.A, Rose, J.P. | | Deposit date: | 2007-04-25 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Altered orientation of active site residues in variants of human ferrochelatase. Evidence for a hydrogen bond network involved in catalysis.
Biochemistry, 46, 2007
|
|
8WZ2
 
 | | Structure of 26RFa-pyroglutamylated RFamide peptide receptor complex | | Descriptor: | G-alpha q, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jin, S, Li, X, Xu, Y, Guo, S, Wu, C, Zhang, H, Yuan, Q, Xu, H.E, Xie, X, Jiang, Y. | | Deposit date: | 2023-11-01 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural basis for recognition of 26RFa by the pyroglutamylated RFamide peptide receptor.
Cell Discov, 10, 2024
|
|
2PNJ
 
 | | Crystal structure of human ferrochelatase mutant with Phe 337 replaced by Ala | | Descriptor: | CHOLIC ACID, FE2/S2 (INORGANIC) CLUSTER, Ferrochelatase, ... | | Authors: | Dailey, H.A, Wu, C.-K, Horanyi, P, Medlock, A.E, Najahi-Missaoui, W, Burden, A.E, Dailey, T.A, Rose, J.P. | | Deposit date: | 2007-04-24 | | Release date: | 2007-10-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Altered orientation of active site residues in variants of human ferrochelatase. Evidence for a hydrogen bond network involved in catalysis
Biochemistry, 46, 2007
|
|
8IWT
 
 | | hSPCA1 in the early E2P state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Calcium-transporting ATPase type 2C member 1, MAGNESIUM ION | | Authors: | Liu, Z.M, Wu, M.Q, Wu, C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structure and transport mechanism of the human calcium pump SPCA1.
Cell Res., 33, 2023
|
|
8IWU
 
 | | hSPCA1 in the E2~P state | | Descriptor: | Calcium-transporting ATPase type 2C member 1, MAGNESIUM ION, TETRAFLUOROALUMINATE ION | | Authors: | Liu, Z.M, Wu, M.Q, Wu, C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structure and transport mechanism of the human calcium pump SPCA1.
Cell Res., 33, 2023
|
|
