6BZY
 
 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the 22D11 broadly neutralizing antibody | | Descriptor: | 22D11 Heavy Chain, 22D11 Light Chain, E2 AS412 peptide | | Authors: | Tzarum, N, Aleman, F, Wilson, I.A, Law, M. | | Deposit date: | 2017-12-26 | | Release date: | 2018-06-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Immunogenetic and structural analysis of a class of HCV broadly neutralizing antibodies and their precursors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2I26
 
 | |
6CA9
 
 | |
6CA7
 
 | |
6BQB
 
 | | MGG4 Fab in complex with peptide | | Descriptor: | GLYCEROL, MGG4 Fab heavy chain, MGG4 Fab light chain, ... | | Authors: | Oyen, D, Tan, J, Lanzavecchia, A, Wilson, I.A. | | Deposit date: | 2017-11-27 | | Release date: | 2018-03-07 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | A public antibody lineage that potently inhibits malaria infection through dual binding to the circumsporozoite protein.
Nat. Med., 24, 2018
|
|
2OK0
 
 | | Fab ED10-DNA complex | | Descriptor: | 5'-D(*TP*C)-3', Fab ED10 heavy chain, Fab ED10 light chain | | Authors: | Stanfield, R.L, Sanguineti, S, Wilson, I.A, de Prat-Gay, G. | | Deposit date: | 2007-01-15 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Specific recognition of a DNA immunogen by its elicited antibody
J.Mol.Biol., 370, 2007
|
|
6BZV
 
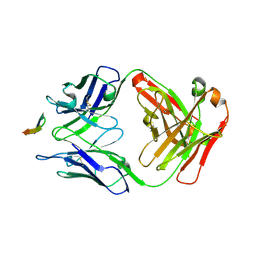 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the GL precursor of the broadly neutralizing antibody 19B3 | | Descriptor: | 19B3 GL Heavy Chain, 19B3 GL Light Chain, E2 AS412 peptide | | Authors: | Tzarum, N, Aleman, F, Wilson, I.A, Law, M. | | Deposit date: | 2017-12-26 | | Release date: | 2018-06-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | Immunogenetic and structural analysis of a class of HCV broadly neutralizing antibodies and their precursors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2O5Z
 
 | |
6BKC
 
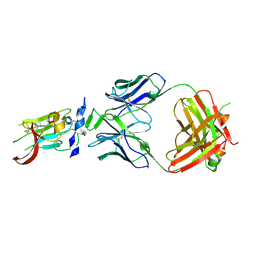 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 6a bound to broadly neutralizing antibody AR3B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab AR3B heavy chain, Fab AR3B light chain, ... | | Authors: | Tzarum, N, Wilson, I.A, Law, M. | | Deposit date: | 2017-11-08 | | Release date: | 2018-12-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Genetic and structural insights into broad neutralization of hepatitis C virus by human VH1-69 antibodies.
Sci Adv, 5, 2019
|
|
6BKO
 
 | |
6BKS
 
 | |
6BL6
 
 | | Crystallization of lipid A transporter MsbA from Salmonella typhimurium | | Descriptor: | Lipid A export ATP-binding/permease protein MsbA | | Authors: | Padayatti, P.S, Stanfield, R.L, Zhang, Q, Wilson, I.A, Lee, S.C. | | Deposit date: | 2017-11-09 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Lipid A Transport Pathway in MsbA.
Structure, 27, 2019
|
|
6BKR
 
 | |
6CA6
 
 | |
6CJK
 
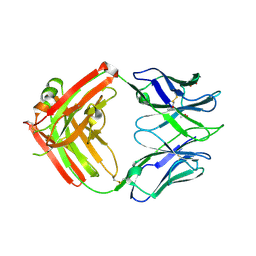 | | Anti HIV Fab 10A | | Descriptor: | ACETATE ION, GLYCEROL, Immunoglobulin Fab heavy chain, ... | | Authors: | Hangartner, L, Ward, A.B, Wilson, I.A, Oyen, D. | | Deposit date: | 2018-02-26 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Electron-Microscopy-Based Epitope Mapping Defines Specificities of Polyclonal Antibodies Elicited during HIV-1 BG505 Envelope Trimer Immunization.
Immunity, 49, 2018
|
|
6CE0
 
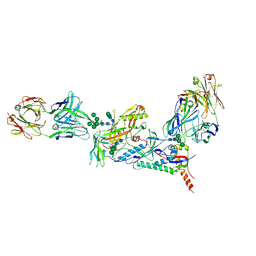 | | Crystal structure of a HIV-1 clade B tier-3 isolate H078.14 UFO-BG Env trimer in complex with broadly neutralizing Fabs PGT124 and 35O22 at 4.6 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 Heavy chain, ... | | Authors: | Kumar, S, Sarkar, A, Wilson, I.A. | | Deposit date: | 2018-02-09 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.602 Å) | | Cite: | HIV-1 vaccine design through minimizing envelope metastability.
Sci Adv, 4, 2018
|
|
6B0N
 
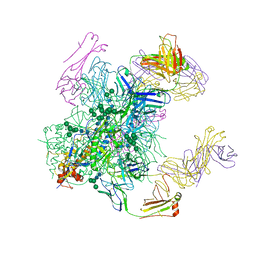 | | Crystal structure of the cleavage-independent prefusion HIV Env glycoprotein trimer of the clade A BG505 isolate (NFL construct) in complex with Fabs PGT122 and PGV19 at 3.39 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp140, ... | | Authors: | Sarkar, A, Irimia, A, Wilson, I.A. | | Deposit date: | 2017-09-14 | | Release date: | 2018-05-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of a cleavage-independent HIV Env recapitulates the glycoprotein architecture of the native cleaved trimer.
Nat Commun, 9, 2018
|
|
2I27
 
 | |
2Q7Y
 
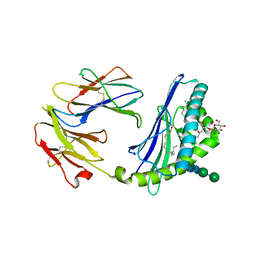 | | Structure of the endogenous iNKT cell ligand iGb3 bound to mCD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Zajonc, D.M, Wilson, I.A, Teyton, L. | | Deposit date: | 2007-06-07 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of Mouse CD1d-iGb3 Complex and its Cognate Valpha14 T Cell Receptor Suggest a Model for Dual Recognition of Foreign and Self Glycolipids.
J.Mol.Biol., 377, 2008
|
|
6AOS
 
 | |
6AOP
 
 | |
6AOQ
 
 | |
2OQJ
 
 | | Crystal structure analysis of Fab 2G12 in complex with peptide 2G12.1 | | Descriptor: | Fab 2G12 heavy chain, Fab 2G12 light chain, peptide 2G12.1 (ACPPSHVLDMRSGTCLAAEGK) | | Authors: | Calarese, D.A, Stanfield, R.L, Menendez, A, Scott, J.K, Wilson, I.A. | | Deposit date: | 2007-01-31 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A peptide inhibitor of HIV-1 neutralizing antibody 2G12 is not a structural mimic of the natural carbohydrate epitope on gp120.
Faseb J., 22, 2008
|
|
2OP4
 
 | | Crystal Structure of Quorum-Quenching Antibody 1G9 | | Descriptor: | 1,2-ETHANEDIOL, Murine Antibody Fab RS2-1G9 IGG1 Heavy Chain, Murine Antibody Fab RS2-1G9 Lambda Light Chain | | Authors: | Kirchdoerfer, R.N, Debler, E.W, Wilson, I.A. | | Deposit date: | 2007-01-26 | | Release date: | 2007-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structures of a Quorum-quenching Antibody.
J.Mol.Biol., 368, 2007
|
|
2JP9
 
 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | Descriptor: | DNA (5'-D(P*DCP*DGP*DCP*DGP*DGP*DGP*DGP*DGP*DCP*DGP*DTP*DCP*DTP*DGP*DCP*DGP*DC)-3'), DNA (5'-D(P*DGP*DCP*DGP*DCP*DAP*DGP*DAP*DCP*DGP*DCP*DCP*DCP*DCP*DCP*DGP*DCP*DG)-3'), Wilms tumor 1, ... | | Authors: | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2007-04-30 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of the wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
