6JYE
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 140K. | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
3IJW
 
 | | Crystal structure of BA2930 in complex with CoA | | Descriptor: | ACETYL COENZYME *A, Aminoglycoside N3-acetyltransferase, CHLORIDE ION, ... | | Authors: | Klimecka, M.M, Chruszcz, M, Skarina, T, Onopryienko, O, Cymborowski, M, Savchenko, A, Edwards, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-05 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
1VK1
 
 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-392566-001 | | Descriptor: | Conserved hypothetical protein, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Shah, A, Liu, Z.J, Tempel, W, Chen, L, Lee, D, Yang, H, Chang, J, Zhao, M, Ng, J, Rose, J, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-04-13 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | (NZ)CH...O contacts assist crystallization of a ParB-like nuclease.
Bmc Struct.Biol., 7, 2007
|
|
1QNJ
 
 | | THE STRUCTURE OF NATIVE PORCINE PANCREATIC ELASTASE AT ATOMIC RESOLUTION (1.1 A) | | Descriptor: | ELASTASE, SODIUM ION, SULFATE ION | | Authors: | Wurtele, M, Hahn, M, Hilpert, K, Hohne, W. | | Deposit date: | 1999-10-15 | | Release date: | 2000-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Structure of Native Porcine Pancreatic Elastase at 1.1 A
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1FVH
 
 | |
3ZUO
 
 | | OMCI in complex with leukotriene B4 | | Descriptor: | COMPLEMENT INHIBITOR, LEUKOTRIENE B4 | | Authors: | Roversi, P, Maillet, I, Togbe, D, Couillin, I, Quesniaux, V.F.J, Teixeira, M, Ahmat, N, Lissina, O, Boland, W, Ploss, K, Caesar, J.J.E, Leonhartsberger, S, Ryffel, B, Lea, S.M, Nunn, M.A. | | Deposit date: | 2011-07-19 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Bifunctional Lipocalin Ameliorates Murine Immune Complex-Induced Acute Lung Injury.
J.Biol.Chem., 288, 2013
|
|
1OHJ
 
 | | HUMAN DIHYDROFOLATE REDUCTASE, MONOCLINIC (P21) CRYSTAL FORM | | Descriptor: | DIHYDROFOLATE REDUCTASE, N-(4-CARBOXY-4-{4-[(2,4-DIAMINO-PTERIDIN-6-YLMETHYL)-AMINO]-BENZOYLAMINO}-BUTYL)-PHTHALAMIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W. | | Deposit date: | 1997-09-17 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Comparison of two independent crystal structures of human dihydrofolate reductase ternary complexes reduced with nicotinamide adenine dinucleotide phosphate and the very tight-binding inhibitor PT523.
Biochemistry, 36, 1997
|
|
1G0R
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). THYMIDINE/GLUCOSE-1-PHOSPHATE COMPLEX. | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION, ... | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-07 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
3ZUI
 
 | | OMCI in complex with palmitoleic acid | | Descriptor: | COMPLEMENT INHIBITOR, PALMITOLEIC ACID | | Authors: | Roversi, P, Maillet, I, Togbe, D, Couillin, I, Quesniaux, V.F.J, Teixeira, M, Ahmat, N, Lissina, O, Boland, W, Ploss, K, Caesar, J.J.E, Leonhartsberger, S, Ryffel, B, Lea, S.M, Nunn, M.A. | | Deposit date: | 2011-07-19 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Bifunctional Lipocalin Ameliorates Murine Immune Complex-Induced Acute Lung Injury.
J.Biol.Chem., 288, 2013
|
|
1OHK
 
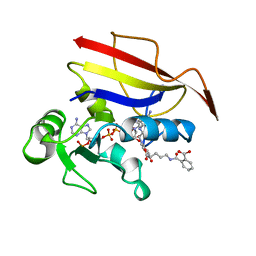 | | HUMAN DIHYDROFOLATE REDUCTASE, ORTHORHOMBIC (P21 21 21) CRYSTAL FORM | | Descriptor: | DIHYDROFOLATE REDUCTASE, N-(4-CARBOXY-4-{4-[(2,4-DIAMINO-PTERIDIN-6-YLMETHYL)-AMINO]-BENZOYLAMINO}-BUTYL)-PHTHALAMIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W. | | Deposit date: | 1997-09-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Comparison of two independent crystal structures of human dihydrofolate reductase ternary complexes reduced with nicotinamide adenine dinucleotide phosphate and the very tight-binding inhibitor PT523.
Biochemistry, 36, 1997
|
|
1FVF
 
 | |
3MW1
 
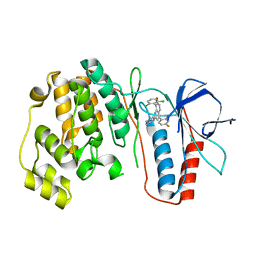 | | p38 kinase Crystal structure in complex with small molecule inhibitor | | Descriptor: | 8-(2,6-dichlorophenyl)-4-(2,4-difluorophenyl)-2-piperidin-4-yl-1,7-naphthyridine 7-oxide, Mitogen-activated protein kinase 14 | | Authors: | Segarra, V, Caturla, F, Lumeras, W, Roca, R, Fisher, M, Lamers, M. | | Deposit date: | 2010-05-05 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 1,7-Naphthyridine 1-Oxides as Novel Potent and Selective Inhibitors of p38 Mitogen Activated Protein Kinase
J.Med.Chem., 54, 2011
|
|
1KD0
 
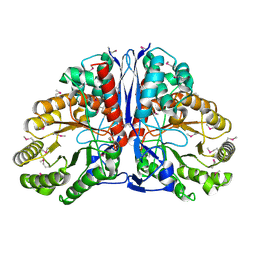 | | Crystal Structure of beta-methylaspartase from Clostridium tetanomorphum. Apo-structure. | | Descriptor: | 1,2-ETHANEDIOL, beta-methylaspartase | | Authors: | Asuncion, M, Blankenfeldt, W, Barlow, J.N, Gani, D, Naismith, J.H. | | Deposit date: | 2001-11-12 | | Release date: | 2001-12-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of 3-methylaspartase from Clostridium tetanomorphum functions via the common enolase chemical step.
J.Biol.Chem., 277, 2002
|
|
3MYG
 
 | | Aurora A Kinase complexed with SCH 1473759 | | Descriptor: | 2-{ethyl[(5-{[6-methyl-3-(1H-pyrazol-4-yl)imidazo[1,2-a]pyrazin-8-yl]amino}isothiazol-3-yl)methyl]amino}-2-methylpropan-1-ol, Serine/threonine-protein kinase 6, TETRAETHYLENE GLYCOL | | Authors: | Hruza, A, Prosis, W, Ramanathan, L. | | Deposit date: | 2010-05-10 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Potent, Injectable Inhibitor of Aurora Kinases Based on the Imidazo-[1,2-a]-Pyrazine Core.
ACS Med Chem Lett, 1, 2010
|
|
1KCZ
 
 | | Crystal Structure of beta-methylaspartase from Clostridium tetanomorphum. Mg-complex. | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, beta-methylaspartase | | Authors: | Asuncion, M, Blankenfeldt, W, Barlow, J.N, Gani, D, Naismith, J.H. | | Deposit date: | 2001-11-12 | | Release date: | 2001-12-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of 3-methylaspartase from Clostridium tetanomorphum functions via the common enolase chemical step.
J.Biol.Chem., 277, 2002
|
|
3MXZ
 
 | | Crystal Structure of tubulin folding cofactor A from Arabidopsis thaliana | | Descriptor: | NITRATE ION, Tubulin-specific chaperone A | | Authors: | Lu, L, Nan, J, Mi, W, Su, X.D, Li, Y. | | Deposit date: | 2010-05-08 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Crystal structure of tubulin folding cofactor A from Arabidopsis thaliana and its beta-tubulin binding characterization
Febs Lett., 584, 2010
|
|
3UGG
 
 | | Crystal structure of a 6-SST/6-SFT from Pachysandra terminalis in complex with 1-kestose | | Descriptor: | GLYCEROL, SULFATE ION, Sucrose:(Sucrose/fructan) 6-fructosyltransferase, ... | | Authors: | Lammens, W, Rabijns, A, Van Laere, A, Strelkov, S.V, Van den Ende, W. | | Deposit date: | 2011-11-02 | | Release date: | 2011-11-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of 6-SST/6-SFT from Pachysandra terminalis, a plant fructan biosynthesizing enzyme in complex with its acceptor substrate 6-kestose.
Plant J., 70, 2012
|
|
4FNV
 
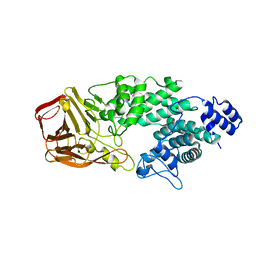 | | Crystal Structure of Heparinase III | | Descriptor: | Heparinase III protein, heparitin sulfate lyase | | Authors: | Dong, W, Ye, S. | | Deposit date: | 2012-06-20 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of heparan sulfate-specific degradation by heparinase III.
Protein Cell, 3, 2012
|
|
6NTS
 
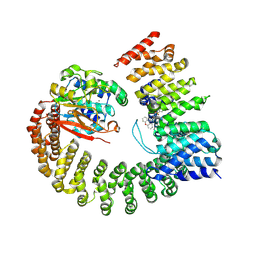 | | Protein Phosphatase 2A (Aalpha-B56alpha-Calpha) holoenzyme in complex with a Small Molecule Activator of PP2A (SMAP) | | Descriptor: | MANGANESE (II) ION, N-[(1R,2R,3S)-2-hydroxy-3-(10H-phenoxazin-10-yl)cyclohexyl]-4-(trifluoromethoxy)benzene-1-sulfonamide, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform, ... | | Authors: | Huang, W, Taylor, D. | | Deposit date: | 2019-01-30 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Selective PP2A Enhancement through Biased Heterotrimer Stabilization.
Cell, 181, 2020
|
|
1KD6
 
 | | Solution structure of the eukaryotic pore-forming cytolysin equinatoxin II | | Descriptor: | EQUINATOXIN II | | Authors: | Hinds, M.G, Zhang, W, Anderluh, G, Hansen, P.E, Norton, R.S. | | Deposit date: | 2001-11-12 | | Release date: | 2002-02-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the eukaryotic pore-forming cytolysin equinatoxin II: implications for pore formation.
J.Mol.Biol., 315, 2002
|
|
1N48
 
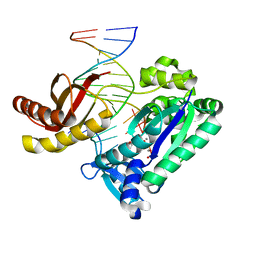 | | Y-family DNA polymerase Dpo4 in complex with DNA containing abasic lesion | | Descriptor: | 5'-D(*CP*AP*(3DR)P*TP*AP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3', 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*CP*TP*A)-3', ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ling, H, Boudsocq, F, Woodgate, R, Yang, W. | | Deposit date: | 2002-10-30 | | Release date: | 2004-02-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Y-family polymerase complexed with abasic lesions: catching DNA with a loaded nucleoside triphosphate
To be Published
|
|
1PVD
 
 | |
4G55
 
 | | Clathrin terminal domain complexed with pitstop 2 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Clathrin heavy chain 1, ... | | Authors: | Bulut, H, Von Kleist, L, Saenger, W, Haucke, V. | | Deposit date: | 2012-07-17 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Role of the clathrin terminal domain in regulating coated pit dynamics revealed by small molecule inhibition.
Cell(Cambridge,Mass.), 146, 2011
|
|
1WAQ
 
 | |
1VPC
 
 | | C-TERMINAL DOMAIN (52-96) OF THE HIV-1 REGULATORY PROTEIN VPR, NMR, 1 STRUCTURE | | Descriptor: | VPR PROTEIN | | Authors: | Schueler, W, De Rocquigny, H, Baudat, Y, Sire, J, Roques, B.P. | | Deposit date: | 1998-02-20 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the (52-96) C-terminal domain of the HIV-1 regulatory protein Vpr: molecular insights into its biological functions.
J.Mol.Biol., 285, 1999
|
|
