5H5R
 
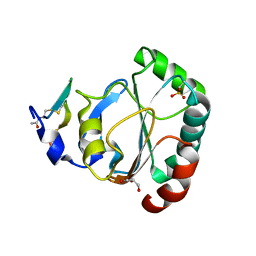 | | Crystal structure of human GPX4 in complex with GXpep-2 | | 分子名称: | GLYCEROL, GXpep-2, Phospholipid hydroperoxide glutathione peroxidase, ... | | 著者 | Sogabe, S, Kadotani, A, Lane, W, Snell, G. | | 登録日 | 2016-11-09 | | 公開日 | 2016-12-07 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Discovery of GPX4 inhibitory peptides from random peptide T7 phage display and subsequent structural analysis
Biochem. Biophys. Res. Commun., 482, 2017
|
|
5H5S
 
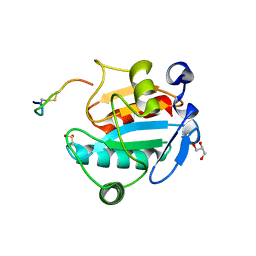 | | Crystal structure of human GPX4 in complex with GXpep-3 | | 分子名称: | GLYCEROL, GXpep-3, Phospholipid hydroperoxide glutathione peroxidase, ... | | 著者 | Sogabe, S, Kadotani, A, Lane, W, Snell, G. | | 登録日 | 2016-11-09 | | 公開日 | 2016-12-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Discovery of GPX4 inhibitory peptides from random peptide T7 phage display and subsequent structural analysis
Biochem. Biophys. Res. Commun., 482, 2017
|
|
7K6E
 
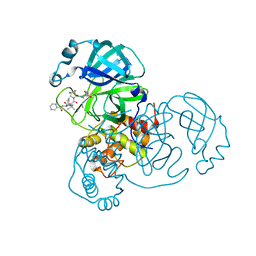 | | SARS-CoV-2 Main Protease Co-Crystal Structure with Telaprevir Determined from Crystals Grown with 40 nL Acoustically Ejected Mpro Droplets at 1.63 A Resolution (Direct Vitrification) | | 分子名称: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Kreitler, D.F, Andi, B, Kumaran, D, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | 登録日 | 2020-09-19 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7K4R
 
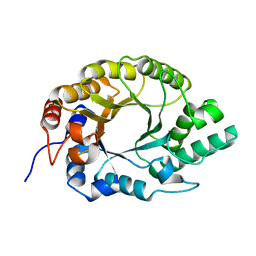 | | Crystal structure of Kemp Eliminase HG3 K50Q | | 分子名称: | Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K4T
 
 | | Crystal structure of Kemp Eliminase HG3.17 | | 分子名称: | Endo-1,4-beta-xylanase | | 著者 | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (0.999 Å) | | 主引用文献 | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
7K5W
 
 | | Cryo-EM structure of heterologous protein complex loaded Thermotoga maritima encapsulin capsid | | 分子名称: | Maritimacin | | 著者 | Xiong, X, Sun, C, Vago, F.S, Klose, T, Zhu, J, Jiang, W. | | 登録日 | 2020-09-17 | | 公開日 | 2020-10-07 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.87 Å) | | 主引用文献 | Cryo-EM Structure of Heterologous Protein Complex Loaded Thermotoga Maritima Encapsulin Capsid.
Biomolecules, 10, 2020
|
|
7TZC
 
 | | A drug and ATP binding site in type 1 ryanodine receptor | | 分子名称: | (2S)-3-(octadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 4-[(7-methoxy-2,3-dihydro-1,4-benzothiazepin-4(5H)-yl)methyl]benzoic acid, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Melville, Z, Dridi, H, Yuan, Q, Reiken, S, Anetta, W, Liu, Y, Clarke, O.B, Marks, A.R. | | 登録日 | 2022-02-15 | | 公開日 | 2022-05-18 | | 最終更新日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (2.45 Å) | | 主引用文献 | A drug and ATP binding site in type 1 ryanodine receptor.
Structure, 30, 2022
|
|
2WFA
 
 | | Structure of Beta-Phosphoglucomutase inhibited with Beryllium trifluoride, in an open conformation. | | 分子名称: | BERYLLIUM TRIFLUORIDE ION, BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION | | 著者 | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | 登録日 | 2009-04-03 | | 公開日 | 2010-05-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Near Attack Conformers Dominate Beta-Phosphoglucomutase Complexes Where Geometry and Charge Distribution Reflect Those of Substrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7K3R
 
 | |
1A11
 
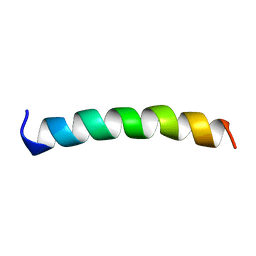 | | NMR STRUCTURE OF MEMBRANE SPANNING SEGMENT 2 OF THE ACETYLCHOLINE RECEPTOR IN DPC MICELLES, 10 STRUCTURES | | 分子名称: | ACETYLCHOLINE RECEPTOR M2 | | 著者 | Gesell, J.J, Sun, W, Montal, M, Opella, S.J. | | 登録日 | 1997-12-19 | | 公開日 | 1998-04-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy.
Nat.Struct.Biol., 6, 1999
|
|
7KED
 
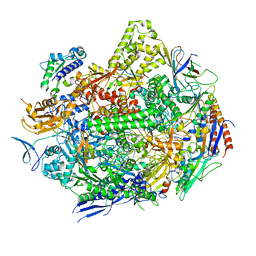 | | RNA polymerase II elongation complex with unnatural base dTPT3 | | 分子名称: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | 著者 | Oh, J, Wang, W, Wang, D. | | 登録日 | 2020-10-10 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Transcriptional processing of an unnatural base pair by eukaryotic RNA polymerase II.
Nat.Chem.Biol., 17, 2021
|
|
2WHE
 
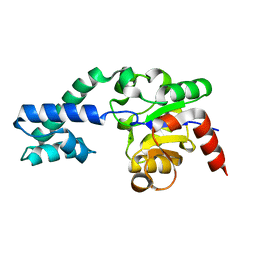 | | Structure of native Beta-Phosphoglucomutase in an open conformation without bound ligands. | | 分子名称: | BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION | | 著者 | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | 登録日 | 2009-05-04 | | 公開日 | 2009-09-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Atomic Details of Near-Transition State Conformers for Enzyme Phosphoryl Transfer Revealed by Mgf-3 Rather Than by Phosphoranes.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
165D
 
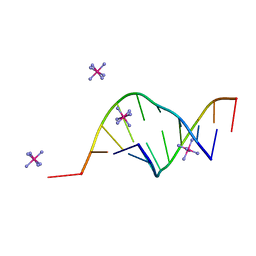 | | THE STRUCTURE OF A MISPAIRED RNA DOUBLE HELIX AT 1.6 ANGSTROMS RESOLUTION AND IMPLICATIONS FOR THE PREDICTION OF RNA SECONDARY STRUCTURE | | 分子名称: | DNA/RNA (5'-R(*GP*CP*UP*UP*CP*GP*GP*CP*)-D(*(BRU))-3'), RHODIUM HEXAMINE ION | | 著者 | Cruse, W, Saludjian, P, Biala, E, Strazewski, P, Prange, T, Kennard, O. | | 登録日 | 1994-03-21 | | 公開日 | 1994-08-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structure of a mispaired RNA double helix at 1.6-A resolution and implications for the prediction of RNA secondary structure.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
2WNC
 
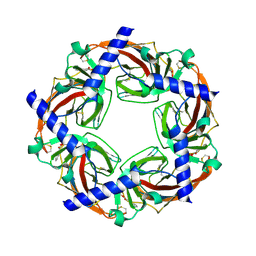 | | Crystal structure of Aplysia ACHBP in complex with tropisetron | | 分子名称: | (3-ENDO)-8-METHYL-8-AZABICYCLO[3.2.1]OCT-3-YL 1H-INDOLE-3-CARBOXYLATE, Soluble acetylcholine receptor | | 著者 | Sulzenbacher, G, Hibbs, R, Shi, J, Talley, T, Conrod, S, Kem, W, Taylor, P, Marchot, P, Bourne, Y. | | 登録日 | 2009-07-08 | | 公開日 | 2009-09-01 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural determinants for interaction of partial agonists with acetylcholine binding protein and neuronal alpha7 nicotinic acetylcholine receptor.
EMBO J., 28, 2009
|
|
7RBY
 
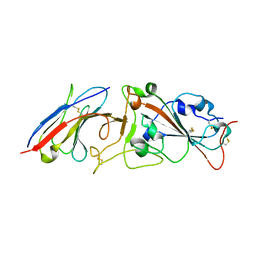 | | Crystal structure of Nanobody nb112 and SARS-CoV-2 RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ilama-isolated nanobody NIH-CoV nb-112 specific to SARS-CoV-2 RBD, MAGNESIUM ION, ... | | 著者 | Chen, Y, Tolbert, W, Pazgier, M. | | 登録日 | 2021-07-06 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.82 Å) | | 主引用文献 | Nebulized delivery of a broadly neutralizing SARS-CoV-2 RBD-specific nanobody prevents clinical, virological, and pathological disease in a Syrian hamster model of COVID-19.
Mabs, 14
|
|
2WF7
 
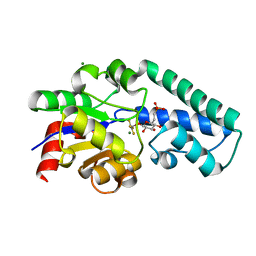 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6- phosphonate and Aluminium tetrafluoride | | 分子名称: | 6,7-dideoxy-7-phosphono-beta-D-gluco-heptopyranose, BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION, ... | | 著者 | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | 登録日 | 2009-04-03 | | 公開日 | 2010-05-19 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Alpha-Fluorophosphonates Reveal How a Phosphomutase Conserves Transition State Conformation Over Hexose Recognition in its Two-Step Reaction.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2WA0
 
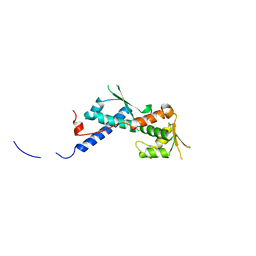 | | Crystal structure of the human MAGEA4 | | 分子名称: | MELANOMA-ASSOCIATED ANTIGEN 4 | | 著者 | Roos, A.K, Cooper, C.D.O, Ugochukwu, E, W Yue, W, Berridge, G, Elkins, J.M, Pike, A.C.W, Bray, J, Filippakopoulos, P, Muniz, J, Chaikuad, A, Burgess-Brown, N, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, von Delft, F, Gileadi, O, Oppermann, U. | | 登録日 | 2009-01-31 | | 公開日 | 2009-03-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structures of Two Melanoma-Associated Antigens Suggest Allosteric Regulation of Effector Binding.
Plos One, 11, 2016
|
|
2WF6
 
 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6-phosphate and Aluminium tetrafluoride | | 分子名称: | 6-O-phosphono-beta-D-glucopyranose, BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION, ... | | 著者 | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | 登録日 | 2009-04-03 | | 公開日 | 2010-05-26 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Atomic details of near-transition state conformers for enzyme phosphoryl transfer revealed by MgF-3 rather than by phosphoranes.
Proc. Natl. Acad. Sci. U.S.A., 107, 2010
|
|
5J39
 
 | | Crystal Structure of the extended TUDOR domain from TDRD2 | | 分子名称: | CACODYLATE ION, Tudor and KH domain-containing protein, UNKNOWN ATOM OR ION | | 著者 | Zhang, H, Tempel, W, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2016-03-30 | | 公開日 | 2016-04-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural basis for arginine methylation-independent recognition of PIWIL1 by TDRD2.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7LMM
 
 | |
7LMK
 
 | |
7LUP
 
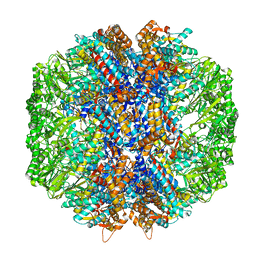 | | Human TRiC/CCT complex with reovirus outer capsid protein sigma-3 | | 分子名称: | Outer capsid protein sigma-3, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | 著者 | Knowlton, J.J, Gestaut, D, Ma, B, Taylor, G, Seven, A.B, Leitner, A, Wilson, G.J, Shanker, S, Yates, N.A, Prasad, B.V.V, Aebersold, R, Chiu, W, Frydman, J, Dermody, T.S. | | 登録日 | 2021-02-22 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (6.2 Å) | | 主引用文献 | Structural and functional dissection of reovirus capsid folding and assembly by the prefoldin-TRiC/CCT chaperone network.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7L9P
 
 | | Structure of human SHLD2-SHLD3-REV7-TRIP13(E253Q) complex | | 分子名称: | Mitotic spindle assembly checkpoint protein MAD2B, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Pachytene checkpoint protein 2 homolog, ... | | 著者 | Xie, W, Patel, D.J. | | 登録日 | 2021-01-04 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Molecular mechanisms of assembly and TRIP13-mediated remodeling of the human Shieldin complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LUM
 
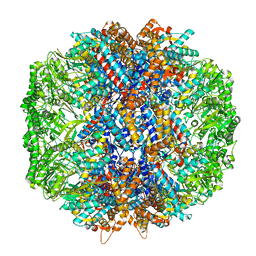 | | Human TRiC in ATP/AlFx closed state | | 分子名称: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | 著者 | Knowlton, J.J, Gestaut, D, Ma, B, Taylor, G, Seven, A.B, Leitner, A, Wilson, G.J, Shanker, S, Yates, N.A, Prasad, B.V.V, Aebersold, R, Chiu, W, Frydman, J, Dermody, T.S. | | 登録日 | 2021-02-22 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structural and functional dissection of reovirus capsid folding and assembly by the prefoldin-TRiC/CCT chaperone network.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LD0
 
 | | Cryo-EM structure of ligand-free Human SARM1 | | 分子名称: | NAD(+) hydrolase SARM1 | | 著者 | Nanson, J.D, Gu, W, Luo, Z, Jia, X, Landsberg, M.J, Kobe, B, Ve, T. | | 登録日 | 2021-01-12 | | 公開日 | 2021-03-10 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | SARM1 is a metabolic sensor activated by an increased NMN/NAD + ratio to trigger axon degeneration.
Neuron, 109, 2021
|
|
