1N4O
 
 | |
3KHE
 
 | | Crystal structure of the calcium-loaded calmodulin-like domain of the CDPK, 541.m00134 from toxoplasma gondii | | Descriptor: | CALCIUM ION, Calmodulin-like domain protein kinase isoform 3, GLYCEROL, ... | | Authors: | Wernimont, A.K, Hutchinson, A, Artz, J.D, Mackenzie, F, Cossar, D, Kozieradzki, I, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Hui, R, Qiu, W, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-30 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of parasitic CDPK domains point to a common mechanism of activation.
Proteins, 79, 2011
|
|
7VVE
 
 | | Complex structure of a leaf-branch compost cutinase variant in complex with mono(2-hydroxyethyl) terephthalic acid | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 4-(2-hydroxyethyloxycarbonyl)benzoic acid, CALCIUM ION, ... | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-05 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
3KZP
 
 | | Crystal structure of putative diguanylate cyclase/phosphodiesterase from Listaria monocytigenes | | Descriptor: | CACODYLATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Klimecka, M.M, Chruszcz, M, Zimmerman, M.D, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-12-08 | | Release date: | 2009-12-22 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative diguanylate cyclase/phosphodiesterase from Listaria monocytigenes
To be Published
|
|
2GLL
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) from Helicobacter pylori | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, BENZAMIDINE, CHLORIDE ION | | Authors: | Zhang, L, Liu, W, Shen, X, Jiang, H. | | Deposit date: | 2006-04-05 | | Release date: | 2007-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ).
J.Biol.Chem., 283, 2008
|
|
3F5L
 
 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, SULFATE ION, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-04 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
7YBN
 
 | |
3KP9
 
 | | Structure of a bacterial homolog of vitamin K epoxide reductase | | Descriptor: | MERCURY (II) ION, UBIQUINONE-10, VKORC1/thioredoxin domain protein | | Authors: | Li, W, Schulman, S, Dutton, R.J, Boyd, D, Beckwith, J, Rapoport, T.A. | | Deposit date: | 2009-11-16 | | Release date: | 2010-02-09 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of a bacterial homologue of vitamin K epoxide reductase.
Nature, 463, 2010
|
|
1N67
 
 | | Clumping Factor A from Staphylococcus aureus | | Descriptor: | Clumping Factor, MAGNESIUM ION | | Authors: | Deivanayagam, C.C.S, Wann, E.R, Chen, W, Carson, M, Rajashankar, K.R, Hook, M, Narayana, S.V.L. | | Deposit date: | 2002-11-08 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel variant of the immunoglobulin fold in surface adhesins of
Staphylococcus aureus: crystal structure of the fibrinogen-binding MSCRAMM,
clumping factor A
Embo J., 21, 2002
|
|
2GPM
 
 | | Crystal structure of an RNA racemate | | Descriptor: | CALCIUM ION, RNA (5'-R(*(0C)P*(0C)P*(0G)P*(0C)P*(0C)P*(0U)P*(0G)P*(0G))-3'), RNA (5'-R(*(0C)P*(0U)P*(0G)P*(0G)P*(0G)P*(0C)P*(0G)P*(0G))-3') | | Authors: | Rypniewski, W, Vallazza, M, Perbandt, M, Klussmann, S, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-04-18 | | Release date: | 2006-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The first crystal structure of an RNA racemate.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
7VVC
 
 | | Crystal structure of inactive mutant of leaf-branch compost cutinase variant | | Descriptor: | ACETATE ION, ACETIC ACID, CALCIUM ION, ... | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-05 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
3FK6
 
 | | Crystal structure of TetR triple mutant (H64K, S135L, S138I) | | Descriptor: | Tetracycline repressor protein class B from transposon Tn10, Tetracycline repressor protein class D | | Authors: | Klieber, M.A, Scholz, O, Lochner, S, Gmeiner, P, Hillen, W, Muller, Y.A. | | Deposit date: | 2008-12-16 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural origins for selectivity and specificity in an engineered bacterial repressor-inducer pair.
Febs J., 276, 2009
|
|
3KH0
 
 | | Crystal structure of the Ras-association (RA) domain of RALGDS | | Descriptor: | Ral guanine nucleotide dissociation stimulator, UNKNOWN ATOM OR ION | | Authors: | Shen, Y, Tempel, W, Wang, H, Tong, Y, Guan, X, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-29 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Ras-association (RA) domain of RALGDS
to be published
|
|
1N6G
 
 | | The structure of immature Dengue-2 prM particles | | Descriptor: | major envelope protein E | | Authors: | Zhang, Y, Corver, J, Chipman, P.R, Zhang, W, Pletnev, S.V, Sedlak, D, Baker, T.S, Strauss, J.H, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2002-11-10 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Structures of Immature flavivirus particles
EMBO J., 22, 2003
|
|
3KE8
 
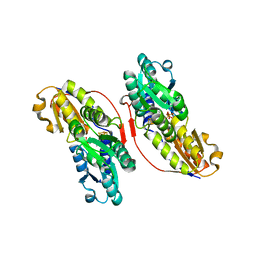 | | Crystal structure of IspH:HMBPP-complex | | Descriptor: | 4-HYDROXY-3-METHYL BUTYL DIPHOSPHATE, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-24 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2H01
 
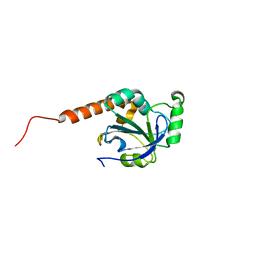 | | PY00414- Plasmodium yoelii thioredoxin peroxidase I | | Descriptor: | 2-CYS PEROXIREDOXIN | | Authors: | Artz, J, Qiu, W, Min, J.R, Dong, A, Lew, J, Melone, M, Alam, Z, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-05-12 | | Release date: | 2006-05-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of PY00414 - a Plasmodium yoelii thioredoxin peroxidase I
To be Published
|
|
2GTA
 
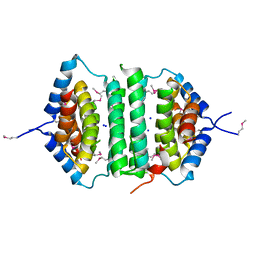 | | Crystal Structure of the putative pyrophosphatase YPJD from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR428. | | Descriptor: | Hypothetical protein ypjD, SODIUM ION | | Authors: | Vorobiev, S.M, Zhou, W, Seetharaman, J, Wang, D, Ma, L.C, Acton, T, Xio, R, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-04-27 | | Release date: | 2006-05-23 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the putative pyrophosphatase YPJD from Bacillus subtilis.
To be Published
|
|
8ABL
 
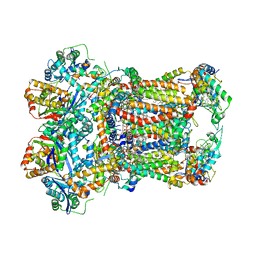 | | Complex III2 from Yarrowia lipolytica, with decylubiquinol and antimycin A, consensus refinement | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, CARDIOLIPIN, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
8ABG
 
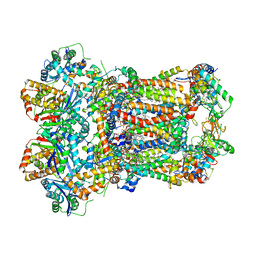 | | Complex III2 from Yarrowia lipolytica, oxidised with ferricyanide, c-position | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, CARDIOLIPIN, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
2GV7
 
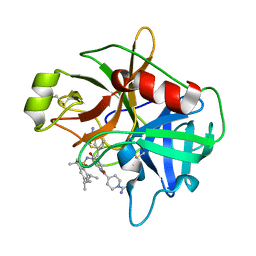 | | Structure of Matriptase in Complex with Inhibitor CJ-672 | | Descriptor: | (S)-4-(4-(3-(3-CARBAMIMIDOYLPHENYL)-2-(2,4,6-TRIISOPROPYLPHENYLSULFONAMIDO)PROPANOYL)PIPERAZINE-1-CARBONYL)PIPERIDINE-1-CARBOXIMIDAMIDE, Suppressor of tumorigenicity 14 | | Authors: | Bode, W. | | Deposit date: | 2006-05-02 | | Release date: | 2006-06-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Secondary Amides of Sulfonylated 3-Amidinophenylalanine. New Potent and Selective Inhibitors of Matriptase.
J.Med.Chem., 49, 2006
|
|
1MWQ
 
 | | Structure of HI0828, a Hypothetical Protein from Haemophilus influenzae with a Putative Active-Site Phosphohistidine | | Descriptor: | CACODYLATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Willis, M.A, Krajewski, W, Chalamasetty, V.R, Reddy, P, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2002-09-30 | | Release date: | 2003-11-25 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Structure of YciI from Haemophilus influenzae (HI0828) reveals a ferredoxin-like alpha/beta-fold with a histidine/aspartate centered catalytic site
Proteins, 59, 2005
|
|
2H2S
 
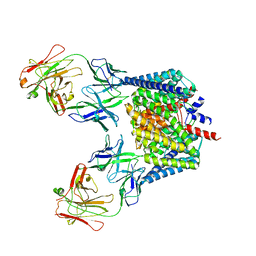 | |
3K19
 
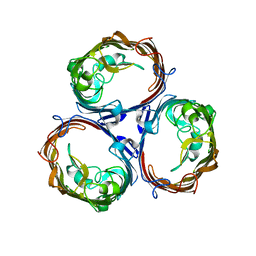 | | OmpF porin | | Descriptor: | Outer membrane protein F | | Authors: | Kefala, G, Ahn, C, Krupa, M, Maslennikov, I, Kwiatkowski, W, Choe, S, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2009-09-26 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | Structures of the OmpF porin crystallized in the presence of foscholine-12.
Protein Sci., 19, 2010
|
|
2H67
 
 | | NMR structure of human insulin mutant HIS-B5-ALA, HIS-B10-ASP PRO-B28-LYS, LYS-B29-PRO, 20 structures | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Hua, Q.X, Liu, M, Hu, S.Q, Jia, W, Arvan, P, Weiss, M.A. | | Deposit date: | 2006-05-30 | | Release date: | 2006-07-18 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | A Conserved Histidine in Insulin Is Required for the Foldability of Human Proinsulin: Structure and function of an Alab5 analog.
J.Biol.Chem., 281, 2006
|
|
3JAE
 
 | | Structure of alpha-1 glycine receptor by single particle electron cryo-microscopy, glycine-bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1 | | Authors: | Du, J, Lu, W, Wu, S.P, Cheng, Y.F, Gouaux, E. | | Deposit date: | 2015-06-08 | | Release date: | 2015-09-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Glycine receptor mechanism elucidated by electron cryo-microscopy.
Nature, 526, 2015
|
|
