4UIE
 
 | | Crystal structure of the S-layer protein SbsC, domains 7, 8 and 9 | | Descriptor: | CALCIUM ION, OSMIUM ION, SURFACE LAYER PROTEIN | | Authors: | Pavkov-Keller, T, Dordic, A, Egelseer, E.M, Sleytr, U.B, Keller, W. | | Deposit date: | 2015-03-27 | | Release date: | 2016-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of the S-Layer Protein Sbsc
To be Published
|
|
4UID
 
 | |
4UIC
 
 | |
5VDB
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with bisubstrate analog 3 | | Descriptor: | (3R,5S,9R,26S)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-10,14,20-trioxo-26-({[(phenylacetyl)amino]acetyl}amino)-2,4,6-trioxa-18-thia-11,15,21-triaza-3,5-diphosphaheptacosan-27-oic acid 3,5-dioxide (non-preferred name), SULFATE ION, acetyltransferase PA4794 | | Authors: | Majorek, K.A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-04-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Generating enzyme and radical-mediated bisubstrates as tools for investigating Gcn5-related N-acetyltransferases.
FEBS Lett., 591, 2017
|
|
6DIG
 
 | | Crystal structure of DQA1*01:02/DQB1*06:02 in complex with a hypocretin peptide | | Descriptor: | 13-mer peptide: ALA-GLY-ASN-HIS-ALA-ALA-GLY-ILE-LEU-THR-LEU-GLY-LYS, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Birtley, J.R, Stern, L.J, Mellins, E.D, Jiang, W. | | Deposit date: | 2018-05-23 | | Release date: | 2019-05-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In vivo clonal expansion and phenotypes of hypocretin-specific CD4+T cells in narcolepsy patients and controls.
Nat Commun, 10, 2019
|
|
4FC4
 
 | | FNT family ion channel | | Descriptor: | Nitrite transporter NirC, octyl beta-D-glucopyranoside | | Authors: | Lue, W, Schwarzer, N, Du, J, Gerbig-Smentek, E, Andrade, S.L.A, Einsle, O. | | Deposit date: | 2012-05-24 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of the nitrite channel NirC from Salmonella typhimurium.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6JM5
 
 | | Crystal structure of TBC1D23 C terminal domain | | Descriptor: | SODIUM ION, TBC1 domain family member 23 | | Authors: | Sun, Q, Huang, W. | | Deposit date: | 2019-03-07 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional studies of TBC1D23 C-terminal domain provide a link between endosomal trafficking and PCH.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4FOX
 
 | | Crystal Structure of Mtb ThyA in complex with dUMP and Raltitrexed | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, TOMUDEX, Thymidylate synthase | | Authors: | Reddy, M.C.M, Bruning, J.B, Sacchettini, J.C, Harshbarger, W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2012-06-21 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of binary and ternary complexes of thymidylate synthase (ThyA) from Mycobacterium tuberculosis: Insights into the selectivity and mode of inhibition
To be Published
|
|
6K7P
 
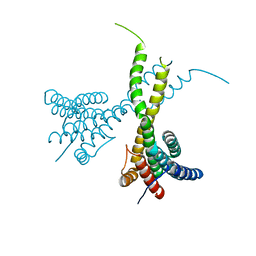 | | Crystal structure of human AFF4-THD domain | | Descriptor: | AF4/FMR2 family member 4 | | Authors: | Tang, D, Xue, Y, Li, S, Cheng, W, Duan, J, Wang, J, Qi, S. | | Deposit date: | 2019-06-08 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional insight into the effect of AFF4 dimerization on activation of HIV-1 proviral transcription.
Cell Discov, 6, 2020
|
|
6YNQ
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2-Methyl-1-tetralone. | | Descriptor: | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6YVF
 
 | | Structure of SARS-CoV-2 Main Protease bound to AZD6482. | | Descriptor: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6WBF
 
 | | Cryo-EM structure of wild type human Pannexin 1 channel | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Lu, W, Du, J, Ruan, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-03 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structures of human pannexin 1 reveal ion pathways and mechanism of gating.
Nature, 584, 2020
|
|
3D0M
 
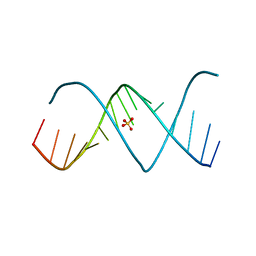 | | X-ray structures of the (GUGGUCUGAUGAGGCC) RNA duplex | | Descriptor: | RNA (5'-R(*GP*UP*GP*GP*UP*CP*UP*GP*AP*UP*GP*AP*GP*GP*CP*C)-3'), SULFATE ION | | Authors: | Rypniewski, W, Adamiak, D.A, Milecki, J, Adamiak, R.W. | | Deposit date: | 2008-05-02 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Noncanonical G(syn)-G(anti) base pairs stabilized by sulphate anions in two X-ray structures of the (GUGGUCUGAUGAGGCC) RNA duplex.
Rna, 14, 2008
|
|
3D18
 
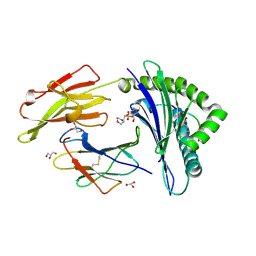 | | Crystal structure of HLA-B*2709 complexed with a variant of the latent membrane protein 2 peptide (LMP2(L)) of epstein-barr virus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-2-microglobulin, ... | | Authors: | Beltrami, A, Gabdulkhakov, A, Rossmann, M, Ziegler, A, Uchanska-Ziegler, B, Saenger, W. | | Deposit date: | 2008-05-05 | | Release date: | 2009-05-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of HLA-B*2709 complexed with a variant of the latent membrane protein 2 peptide (LMP2(L)) of epstein-barr virus
To be Published
|
|
3D3T
 
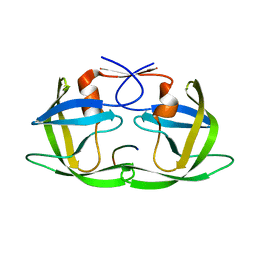 | | Crystal Structure of HIV-1 CRF01_AE in complex with the substrate p1-p6 | | Descriptor: | HIV-1 protease, P1-P6 SUBSTRATE PEPTIDE | | Authors: | Bandaranayake, R.M, Prabu-Jeyabalan, M, Kakizawa, J, Sugiura, W, Schiffer, C.A. | | Deposit date: | 2008-05-12 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of human immunodeficiency virus type 1 CRF01_AE protease in complex with the substrate p1-p6.
J.Virol., 82, 2008
|
|
3D4B
 
 | | Crystal structure of Sir2Tm in complex with Acetyl p53 peptide and DADMe-NAD+ | | Descriptor: | 5'-O-[(R)-{[(R)-{[(3R,4R)-1-(3-carbamoylbenzyl)-4-hydroxypyrrolidin-3-yl]methoxy}(hydroxy)phosphoryl]methyl}(hydroxy)phosphoryl]adenosine, Acetyl P53 peptide, NAD-dependent deacetylase, ... | | Authors: | Hawse, W.F, Hoff, K.G, Fatkins, D, Daines, A, Zubkova, O.V, Schramm, V.L, Zheng, W, Wolberger, C. | | Deposit date: | 2008-05-14 | | Release date: | 2008-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into intermediate steps in the Sir2 deacetylation reaction.
Structure, 16, 2008
|
|
6W2E
 
 | | Structures of Capsid and Capsid-Associated Tegument Complex inside the Epstein-Barr Virus | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Liu, W, Cui, Y.X, Wang, C.Y, Li, Z.H, Gong, D.Y, Dai, X.H, Bi, G.Q, Sun, R, Zhou, Z.H. | | Deposit date: | 2020-03-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structures of capsid and capsid-associated tegument complex inside the Epstein-Barr virus.
Nat Microbiol, 5, 2020
|
|
6W10
 
 | |
6VQP
 
 | | Structure of CalU17 from the Calicheamicin Biosynthesis Pathway of Micromonospora echinospora | | Descriptor: | CalU17, CalU17 His-Tagged protein, GLYCEROL, ... | | Authors: | Kosgei, A.J, Miller, M.D, Xu, W, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2020-02-05 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of DynF from the Dynemicin Biosynthesis Pathway of Micromonospora chersina
To Be Published
|
|
3CEZ
 
 | | Crystal structure of methionine-R-sulfoxide reductase from Burkholderia pseudomallei | | Descriptor: | ACETIC ACID, Methionine-R-sulfoxide reductase, ZINC ION | | Authors: | Staker, B, Napuli, A, Nakazawa, S.H, Castaneda, L, Alkafeef, S, Vanvoorhis, W, Stewart, L, Myler, P, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2008-02-29 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methionine-R-sulfoxide reductase from Burkholderia pseudomallei.
To be Published
|
|
3CWC
 
 | | Crystal structure of putative glycerate kinase 2 from Salmonella typhimurium LT2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative glycerate kinase 2 | | Authors: | Osipiuk, J, Zhou, M, Holzle, D, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-04-21 | | Release date: | 2008-07-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | X-ray crystal structure of putative glycerate kinase 2 from Salmonella typhimurium LT2.
To be Published
|
|
3CNV
 
 | | Crystal structure of the ligand-binding domain of a putative GntR-family transcriptional regulator from Bordetella bronchiseptica | | Descriptor: | CHLORIDE ION, CITRATE ANION, Putative GntR-family transcriptional regulator | | Authors: | Zimmerman, M.D, Xu, X, Cui, H, Filippova, E.V, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-26 | | Release date: | 2008-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the ligand-binding domain of a putative GntR-family transcriptional regulator from Bordetella bronchiseptica.
To be Published
|
|
1BJZ
 
 | |
3D26
 
 | | Norwalk P domain A-trisaccharide complex | | Descriptor: | 58 kd capsid protein, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)]beta-D-galactopyranose | | Authors: | Hegde, R, Bu, W. | | Deposit date: | 2008-05-07 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the receptor binding specificity of Norwalk virus.
J.Virol., 82, 2008
|
|
6WVE
 
 | | Chicken SPCS1 | | Descriptor: | Green fluorescent protein,Chicken Signal Peptidase Complex Subunit 1,Green fluorescent protein | | Authors: | Liu, S, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2021-01-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.429 Å) | | Cite: | Termini restraining of small membrane proteins enables structure determination at near-atomic resolution.
Sci Adv, 6, 2020
|
|
