7FIW
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidAwMel(ST) and CidBND1-ND2 from wPip(Pel) | | 分子名称: | ULP_PROTEASE domain-containing protein, bacteria factor 4,CidA I(Zeta/1) protein | | 著者 | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-08-01 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
5H1B
 
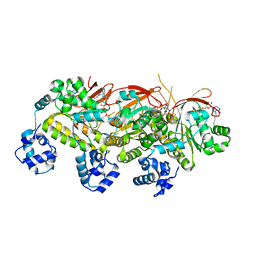 | | Human RAD51 presynaptic complex | | 分子名称: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA repair protein RAD51 homolog 1, MAGNESIUM ION, ... | | 著者 | Xu, J, Zhao, L, Xu, Y, Zhao, W, Sung, P, Wang, H.W. | | 登録日 | 2016-10-08 | | 公開日 | 2016-12-21 | | 最終更新日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Cryo-EM structures of human RAD51 recombinase filaments during catalysis of DNA-strand exchange
Nat. Struct. Mol. Biol., 24, 2017
|
|
7FIU
 
 | | Crystal structure of the DUB domain of Wolbachia cytoplasmic incompatibility factor CidB from wMel | | 分子名称: | ULP_PROTEASE domain-containing protein | | 著者 | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-08-01 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIT
 
 | | Crystal structure of Wolbachia cytoplasmic incompatibility factor CidA from wMel | | 分子名称: | bacteria factor 1 | | 著者 | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-08-01 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
6JYA
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with CW laser (ND-10%) at 95K. | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | 登録日 | 2019-04-26 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.803 Å) | | 主引用文献 | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
5GNX
 
 | | The E171Q mutant structure of Bgl6 | | 分子名称: | Beta-glucosidase, GLYCEROL, PROPANOIC ACID, ... | | 著者 | Xie, W, Pang, P, Cao, L.C, Liu, Y.H, Wang, Z. | | 登録日 | 2016-07-25 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structures of a glucose-tolerant beta-glucosidase provide insights into its mechanism.
J. Struct. Biol., 198, 2017
|
|
5EMF
 
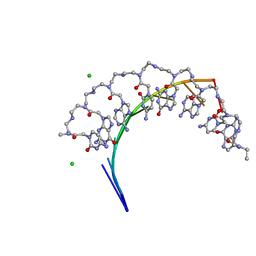 | | Crystal structure of RNA r(GCUGCUGC) with antisense PNA p(GCAGCAGC) | | 分子名称: | CHLORIDE ION, RNA (5'-R(*GP*CP*UP*GP*CP*UP*GP*C)-3'), antisense PNA p(GCAGCAGC) | | 著者 | Kiliszek, A, Banaszak, K, Dauter, Z, Rypniewski, W. | | 登録日 | 2015-11-06 | | 公開日 | 2016-01-13 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.14 Å) | | 主引用文献 | The first crystal structures of RNA-PNA duplexes and a PNA-PNA duplex containing mismatches-toward anti-sense therapy against TREDs.
Nucleic Acids Res., 44, 2016
|
|
2PPZ
 
 | |
6JYF
 
 | | Structure of light-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 140K. | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | 登録日 | 2019-04-26 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.004 Å) | | 主引用文献 | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
2PUT
 
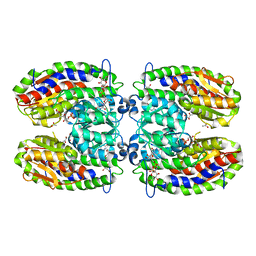 | | The crystal structure of isomerase domain of glucosamine-6-phosphate synthase from Candida albicans | | 分子名称: | ACETATE ION, FRUCTOSE -6-PHOSPHATE, SODIUM ION, ... | | 著者 | Raczynska, J, Olchowy, J, Milewski, S, Rypniewski, W. | | 登録日 | 2007-05-09 | | 公開日 | 2007-09-11 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The Crystal and Solution Studies of Glucosamine-6-phosphate Synthase from Candida albicans
J.Mol.Biol., 372, 2007
|
|
7ESH
 
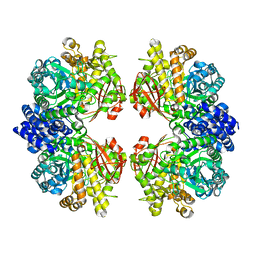 | | Crystal structure of amylosucrase from Calidithermus timidus | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, amylosucrase | | 著者 | Tian, Y, Hou, X, Ni, D, Xu, W, Guang, C, Zhang, W, Rao, Y, Mu, W. | | 登録日 | 2021-05-10 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Structure-based interface engineering methodology in designing a thermostable amylose-forming transglucosylase
J.Biol.Chem., 298, 2022
|
|
1V5D
 
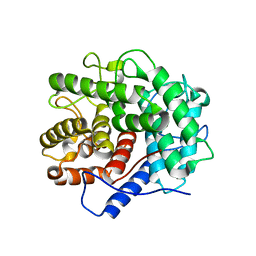 | | The crystal structure of the active form chitosanase from Bacillus sp. K17 at pH6.4 | | 分子名称: | PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), chitosanase | | 著者 | Adachi, W, Shimizu, S, Sunami, T, Fukazawa, T, Suzuki, M, Yatsunami, R, Nakamura, S, Takenaka, A. | | 登録日 | 2003-11-22 | | 公開日 | 2004-12-07 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of family GH-8 chitosanase with subclass II specificity from Bacillus sp. K17
J.MOL.BIOL., 343, 2004
|
|
3OG4
 
 | | The crystal structure of human interferon lambda 1 complexed with its high affinity receptor in space group P21212 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin 28 receptor, alpha (Interferon, ... | | 著者 | Miknis, Z.J, Magracheva, E, Lei, W, Zdanov, A, Kotenko, S.V, Wlodawer, A. | | 登録日 | 2010-08-16 | | 公開日 | 2010-10-20 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Crystal structure of the complex of human interferon-lambda1 with its high affinity receptor interferon-lambdaR1.
J.Mol.Biol., 404, 2010
|
|
5GRJ
 
 | | Crystal structure of human PD-L1 with monoclonal antibody avelumab | | 分子名称: | Programmed cell death 1 ligand 1, avelumab H chain, avelumab L chain | | 著者 | Liu, K, Tan, S, Chai, Y, Chen, D, Song, H, Zhang, C.W.-H, Shi, Y, Liu, J, Tan, W, Lyu, J, Gao, S, Yan, J, Qi, J, Gao, G.F. | | 登録日 | 2016-08-11 | | 公開日 | 2016-11-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.206 Å) | | 主引用文献 | Structural basis of anti-PD-L1 monoclonal antibody avelumab for tumor therapy.
Cell Res., 27, 2017
|
|
5GSV
 
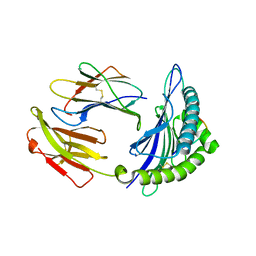 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 142-5 | | 分子名称: | 10-mer peptide from Spike protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | 著者 | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | 登録日 | 2016-08-17 | | 公開日 | 2017-04-26 | | 実験手法 | X-RAY DIFFRACTION (1.996 Å) | | 主引用文献 | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5GNZ
 
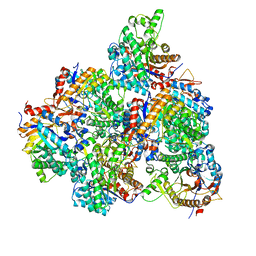 | | The M3 mutant structure of Bgl6 | | 分子名称: | Beta-glucosidase, GLYCEROL, beta-D-glucopyranose | | 著者 | Xie, W, Pang, P, Cao, L.C, Liu, Y.H, Wang, Z. | | 登録日 | 2016-07-25 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structures of a glucose-tolerant beta-glucosidase provide insights into its mechanism.
J. Struct. Biol., 198, 2017
|
|
5H5F
 
 | |
5GSR
 
 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide I5A | | 分子名称: | 9-mer peptide from Spike protein, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | 著者 | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | 登録日 | 2016-08-17 | | 公開日 | 2017-04-26 | | 実験手法 | X-RAY DIFFRACTION (2.198 Å) | | 主引用文献 | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
3OG6
 
 | | The crystal structure of human interferon lambda 1 complexed with its high affinity receptor in space group P212121 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Interleukin 28 receptor, ... | | 著者 | Miknis, Z.J, Magracheva, E, Lei, W, Zdanov, A, Kotenko, S.V, Wlodawer, A. | | 登録日 | 2010-08-16 | | 公開日 | 2010-10-20 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.097 Å) | | 主引用文献 | Crystal structure of the complex of human interferon-lambda1 with its high affinity receptor interferon-lambdaR1.
J.Mol.Biol., 404, 2010
|
|
5GUF
 
 | |
5GR7
 
 | | Mouse MHC class I H-2Kd with a MERS-CoV-derived peptide 37-1 | | 分子名称: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-D alpha chain, ... | | 著者 | Liu, K, Chai, Y, Qi, J, Tan, W, Liu, W.J, Gao, G.F. | | 登録日 | 2016-08-08 | | 公開日 | 2017-06-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Protective T Cell Responses Featured by Concordant Recognition of Middle East Respiratory Syndrome Coronavirus-Derived CD8+ T Cell Epitopes and Host MHC.
J. Immunol., 198, 2017
|
|
5GWD
 
 | | Structure of Myroilysin | | 分子名称: | Myroilysin, ZINC ION | | 著者 | Xu, D, Ran, T, Wang, W. | | 登録日 | 2016-09-10 | | 公開日 | 2017-02-15 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Myroilysin Is a New Bacterial Member of the M12A Family of Metzincin Metallopeptidases and Is Activated by a Cysteine Switch Mechanism.
J. Biol. Chem., 292, 2017
|
|
5H1C
 
 | | Human RAD51 post-synaptic complexes | | 分子名称: | DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | 著者 | Xu, J, Zhao, L, Xu, Y, Zhao, W, Sung, P, Wang, H.W. | | 登録日 | 2016-10-08 | | 公開日 | 2016-12-21 | | 最終更新日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Cryo-EM structures of human RAD51 recombinase filaments during catalysis of DNA-strand exchange
Nat. Struct. Mol. Biol., 24, 2017
|
|
6JYE
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 140K. | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | 登録日 | 2019-04-26 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
5GX6
 
 | | Crystal structure of solute-binding protein complexed with unsaturated chondroitin disaccharide with a sulfate group at C-4 position of GalNAc | | 分子名称: | 1,2-ETHANEDIOL, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, CALCIUM ION, ... | | 著者 | Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | 登録日 | 2016-09-15 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | A bacterial ABC transporter enables import of mammalian host glycosaminoglycans
Sci Rep, 7, 2017
|
|
