7YKO
 
 | | Crystal structure of a novel alpha/beta hydrolase mutant from thermomonospora curvata in complex with pentane-1,5-diol | | Descriptor: | Triacylglycerol lipase, pentane-1,5-diol | | Authors: | Han, X, Jian, G, Bornscheuer, U.T, Wei, R, Liu, W. | | Deposit date: | 2022-07-23 | | Release date: | 2023-07-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of a novel alpha/beta hydrolase mutant from thermomonospora curvata in complex with pentane-1,5-diol
To Be Published
|
|
7YKQ
 
 | |
7YLQ
 
 | | Crystal structure of Microcystinase C from Sphingomonas sp. ACM-3962 at 2.6 A resolution | | Descriptor: | CALCIUM ION, Microcystinase C, ZINC ION | | Authors: | Guo, X, Li, Z, Ding, W, Yin, P, Feng, L. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of Microcystin C from Sphingomonas sp. ACM-3962 at 2.6 A resolution
To Be Published
|
|
7YMB
 
 | |
7YMW
 
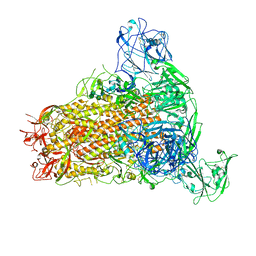 | | Cryo-EM structure of MERS-CoV spike protein, One RBD-up conformation 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (6.05 Å) | | Cite: | Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries.
Cell, 187, 2024
|
|
7YMX
 
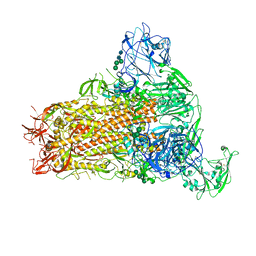 | | Cryo-EM structure of MERS-CoV spike protein, One RBD-up conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.44 Å) | | Cite: | Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries.
Cell, 187, 2024
|
|
7YMT
 
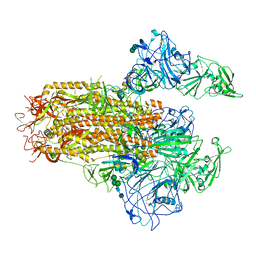 | | Cryo-EM structure of MERS-CoV spike protein, Two RBD-up conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (6.55 Å) | | Cite: | Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries.
Cell, 187, 2024
|
|
7YN0
 
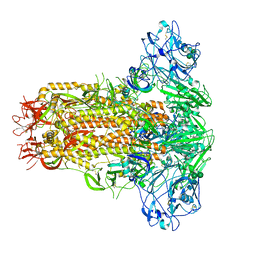 | | Cryo-EM structure of MERS-CoV spike protein, all RBD-down conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries.
Cell, 187, 2024
|
|
7YMY
 
 | | Cryo-EM structure of MERS-CoV spike protein, One RBD-up conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Hsu, S.T.D, Chang, N.E, Weng, Z.W, Yang, T.J, Draczkowski, P. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-09 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.96 Å) | | Cite: | Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries.
Cell, 187, 2024
|
|
5HVT
 
 | | Crystal Structure of Macrophage Migration Inhibitory Factor (MIF) with a Potent Inhibitor (NVS-2) | | Descriptor: | 7-hydroxy-3-(4-methoxyphenyl)-3,4-dihydro-2H-1,3-benzoxazin-2-one, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Robertson, M.J, Jorgensen, W.L. | | Deposit date: | 2016-01-28 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Fluorescence Polarization Assay for Binding to Macrophage Migration Inhibitory Factor and Crystal Structures for Complexes of Two Potent Inhibitors.
J.Am.Chem.Soc., 138, 2016
|
|
7YRS
 
 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI complexed with MOPS | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase, ZINC ION | | Authors: | Yamamoto, Y, Oiki, S, Takase, R, Mikami, B, Hashimoto, W. | | Deposit date: | 2022-08-10 | | Release date: | 2023-08-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Crystal Structures of Lacticaseibacillus 4-Deoxy-L- threo- 5-hexosulose-uronate Ketol-isomerase KduI in Complex with Substrate Analogs.
J Appl Glycosci (1999), 70, 2023
|
|
7YPL
 
 | |
7ARK
 
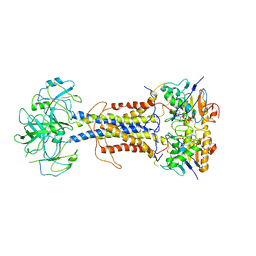 | | LolCDE in complex with AMP-PNP in the closed NBD state | | Descriptor: | Lipoprotein-releasing ABC transporter permease subunit LolC, Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolE, ... | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7B73
 
 | | Insight into the molecular determinants of thermal stability in halohydrin dehalogenase HheD2. | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Short-chain dehydrogenase/reductase SDR | | Authors: | Wessel, J, Petrillo, G, Estevez, M, Bosh, S, Seeger, M, Dijkman, W.P, Hidalgo, A, Uson, I, Osuna, S, Schallmey, A. | | Deposit date: | 2020-12-09 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into the molecular determinants of thermal stability in halohydrin dehalogenase HheD2.
Febs J., 288, 2021
|
|
7ARJ
 
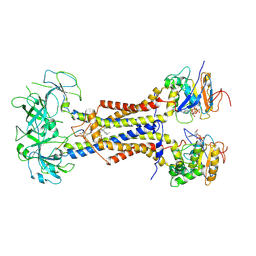 | | LolCDE in complex with lipoprotein and AMPPNP complex undimerized form | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, LPP, Lipoprotein-releasing ABC transporter permease subunit LolC, ... | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7YW1
 
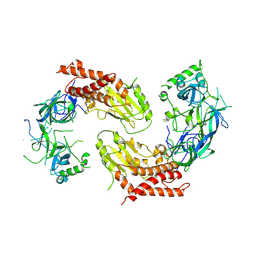 | | crystal structure of UBE2O | | Descriptor: | (E3-independent) E2 ubiquitin-conjugating enzyme UBE2O | | Authors: | Fu, Z, Zhu, W, Huang, H. | | Deposit date: | 2022-08-20 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | crystal structure of UBE2O
To Be Published
|
|
7ARI
 
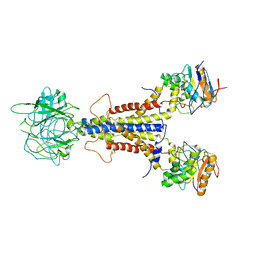 | | LolCDE apo structure | | Descriptor: | Lipoprotein-releasing ABC transporter permease subunit LolC, Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolE | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7ARH
 
 | | LolCDE in complex with lipoprotein | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, LPP, Lipoprotein-releasing ABC transporter permease subunit LolC, ... | | Authors: | Tang, X.D, Chang, S.H, Zhang, K, Wang, T, Luo, Q.H, Qiao, W, Wang, C, Zhang, Z.B, Zhang, Z.Y, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2020-10-25 | | Release date: | 2021-04-07 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for bacterial lipoprotein relocation by the transporter LolCDE.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6PZO
 
 | |
6PZU
 
 | |
6Q06
 
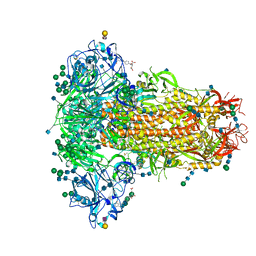 | | MERS-CoV S structure in complex with 2,3-sialyl-N-acetyl-lactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Park, Y.J, Walls, A.C, Wang, Z, Sauer, M, Li, W, Tortorici, M.A, Bosch, B.J, DiMaio, F.D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7YRR
 
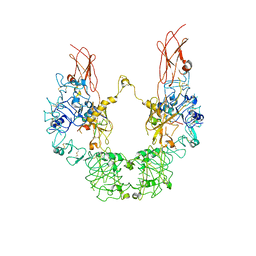 | | Cryo-EM structure of IGF1R with two IGF1 complex | | Descriptor: | Insulin-like growth factor 1 receptor, Isoform 3 of Insulin-like growth factor I | | Authors: | Xi, Z, Cang, W. | | Deposit date: | 2022-08-10 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of IGF1R with two IGF1 complex at 4.3 angstroms resolution
To Be Published
|
|
6Q4Y
 
 | | Structure of MPT-2, a GDP-Man-dependent mannosyltransferase from Leishmania mexicana, in complex with mannose | | Descriptor: | LmxM MPT-2, alpha-D-mannopyranose, beta-D-mannopyranose | | Authors: | Sobala, L.F, Males, A, Bastidas, L.M, Ward, T, Sernee, M.F, Ralton, J.E, Nero, T.L, Cobbold, S, Kloehn, J, Viera-Lara, M, Stanton, L, Hanssen, E, Parker, M.W, Williams, S.J, McConville, M.J, Davies, G.J. | | Deposit date: | 2018-12-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Family of Dual-Activity Glycosyltransferase-Phosphorylases Mediates Mannogen Turnover and Virulence in Leishmania Parasites.
Cell Host Microbe, 26, 2019
|
|
6Q4X
 
 | | Structure of MPT-2, a GDP-Man-dependent mannosyltransferase from Leishmania mexicana | | Descriptor: | SODIUM ION, Uncharacterized protein | | Authors: | Sobala, L.F, Males, A, Bastidas, L.M, Ward, T, Sernee, M.F, Ralton, J.E, Nero, T.L, Cobbold, S, Kloehn, J, Viera-Lara, M, Stanton, L, Hanssen, E, Parker, M.W, Williams, S.J, McConville, M.J, Davies, G.J. | | Deposit date: | 2018-12-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Family of Dual-Activity Glycosyltransferase-Phosphorylases Mediates Mannogen Turnover and Virulence in Leishmania Parasites.
Cell Host Microbe, 26, 2019
|
|
5HNF
 
 | | Crystal structure of pyrene- and phenanthrene-modified DNA in complex with the BpuJ1 endonuclease binding domain | | Descriptor: | DNA (5'-D(*GP*(YPE)P*AP*CP*CP*CP*GP*TP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*AP*CP*GP*GP*GP*TP*(YPF)P*C)-3'), Restriction endonuclease R.BpuJI | | Authors: | Probst, M, Aeschimann, W, Chau, T.-T.-H, Langenegger, S.M, Stocker, A, Haener, R. | | Deposit date: | 2016-01-18 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.546 Å) | | Cite: | Structural insight into DNA-assembled oligochromophores: crystallographic analysis of pyrene- and phenanthrene-modified DNA in complex with BpuJI endonuclease.
Nucleic Acids Res., 44, 2016
|
|
