4V5X
 
 | | The cryo-EM structure of a 3D DNA-origami object | | Descriptor: | SCAFFOLD STRAND,SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Bai, X.C, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2012-10-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Cryo-Em Structure of a 3D DNA-Origami Object.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4V54
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with ribosome recycling factor (RRF). | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | Deposit date: | 2007-06-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
4V1O
 
 | | Architecture of the RNA polymerase II-Mediator core transcription initiation complex | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Plaschka, C, Lariviere, L, Wenzeck, L, Hemann, M, Tegunov, D, Petrotchenko, E.V, Borchers, C.H, Baumeister, W, Herzog, F, Villa, E, Cramer, P. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Architecture of the RNA Polymerase II-Mediator Core Initiation Complex.
Nature, 518, 2015
|
|
4V4L
 
 | | Structure of the Drosophila apoptosome | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apaf-1 related killer DARK, MAGNESIUM ION | | Authors: | Yuan, S, Topf, M, Akey, C.W, Ludtke, S.J. | | Deposit date: | 2010-10-04 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structure of the Drosophila apoptosome at 6.9 angstrom resolution
Structure, 19, 2011
|
|
4V5M
 
 | | tRNA tranlocation on the 70S ribosome: the pre-translocational translocation intermediate TI(PRE) | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ratje, A.H, Loerke, J, Mikolajka, A, Bruenner, M, Hildebrand, P.W, Starosta, A.L, Doenhoefer, A, Connell, S.R, Fucini, P, Mielke, T, Whitford, P.C, Onuchic, J.N, Yu, Y, Sanbonmatsu, K.Y, Hartmann, R.K, Penczek, P.A, Wilson, D.N, Spahn, C.M.T. | | Deposit date: | 2010-10-01 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Head Swivel on the Ribosome Facilitates Translocation by Means of Intra-Subunit tRNA Hybrid Sites.
Nature, 468, 2010
|
|
4V9G
 
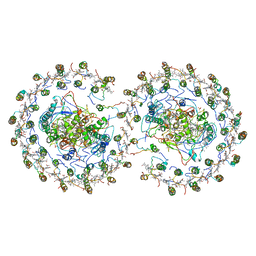 | | RC-LH1-PufX dimer complex from Rhodobacter sphaeroides | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (II) ION, ... | | Authors: | Qian, P, Papiz, M.Z, Jackson, P.J, Brindley, A.A, Ng, I.W, Olsen, J.D, Dickman, M.J, Bullough, P.A, Hunter, C.N. | | Deposit date: | 2013-02-21 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (7.78 Å) | | Cite: | Three-Dimensional Structure of the Rhodobacter sphaeroides RC-LH1-PufX Complex: Dimerization and Quinone Channels Promoted by PufX.
Biochemistry, 52, 2013
|
|
4V92
 
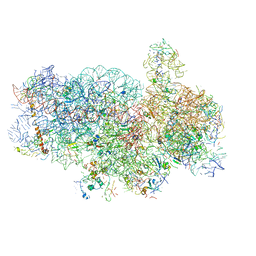 | | Kluyveromyces lactis 80S ribosome in complex with CrPV-IRES | | Descriptor: | 18S RRNA, ES1, ES10, ... | | Authors: | Fernandez, I.S, Bai, X, Scheres, S.H.W, Ramakrishnan, V. | | Deposit date: | 2014-03-21 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Initiation of Translation by Cricket Paralysis Virus Ires Requires its Translocation in the Ribosome.
Cell(Cambridge,Mass.), 157, 2014
|
|
4UW1
 
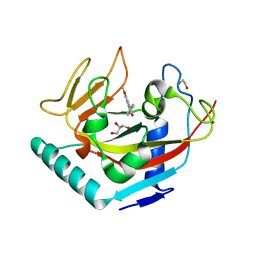 | | X-ray crystal structure of human TNKS in complex with a small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-{4-[(dimethylamino)methyl]phenyl}-5-methoxyisoquinolin-1(2H)-one, GLYCEROL, ... | | Authors: | Oliver, A.W, Rajasekaran, M.B, Pearl, L.H. | | Deposit date: | 2014-08-08 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Design and Discovery of 3-Aryl-5-Substituted-Isoquinolin-1-Ones as Potent and Selective Tankyrase Inhibitors
Medchemcommm, 6, 2015
|
|
4URE
 
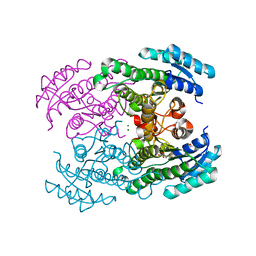 | | Molecular Genetic and Crystal Structural Analysis of 1-(4- Hydroxyphenyl)-Ethanol Dehydrogenase from Aromatoleum aromaticum EbN1 | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ACETATE ION, CYCLOHEXANOL DEHYDROGENASE, ... | | Authors: | Buesing, I, Hoeffken, H.W, Breuer, M, Woehlbrand, L, Hauer, B, Rabus, R. | | Deposit date: | 2014-06-28 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Genetic and Crystal Structural Analysis of 1-(4-Hydroxyphenyl)-Ethanol Dehydrogenase from 'Aromatoleum Aromaticum' Ebn1.
J.Mol.Microbiol.Biotechnol., 25, 2015
|
|
4V1N
 
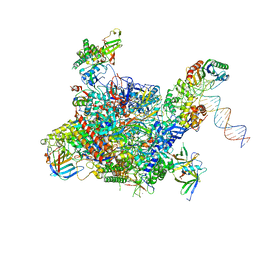 | | Architecture of the RNA polymerase II-Mediator core transcription initiation complex | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Plaschka, C, Lariviere, L, Wenzeck, L, Hemann, M, Tegunov, D, Petrotchenko, E.V, Borchers, C.H, Baumeister, W, Herzog, F, Villa, E, Cramer, P. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Architecture of the RNA Polymerase II-Mediator Core Initiation Complex.
Nature, 518, 2015
|
|
6WHM
 
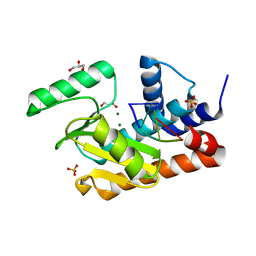 | | The crystal structure of the 2009/H1N1/California PA endonuclease wild type bound to DNA oligomer TAGC (cleaved TTAGCATT, 5mM overnight DNA soak) | | Descriptor: | DNA (5'-D(P*TP*AP*GP*C)-3'), GLYCEROL, MAGNESIUM ION, ... | | Authors: | Cuypers, M.G, Kumar, G, Webb, T, White, S.W. | | Deposit date: | 2020-04-08 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural insights into the substrate specificity of the endonuclease activity of the influenza virus cap-snatching mechanism.
Nucleic Acids Res., 49, 2021
|
|
5KIY
 
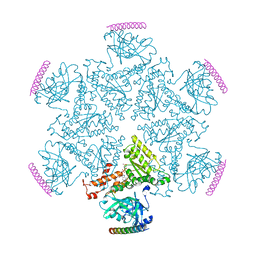 | | p97 ND1-A232E in complex with VIMP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Selenoprotein S, ... | | Authors: | Tang, W.K, Xia, D. | | Deposit date: | 2016-06-17 | | Release date: | 2017-12-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis for nucleotide-modulated p97 association with the ER membrane.
Cell Discov, 3, 2017
|
|
5KRH
 
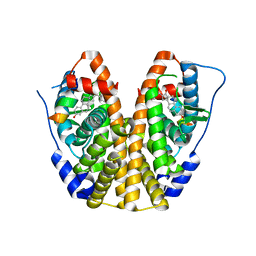 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 16-benzylidene estrone | | Descriptor: | (8~{R},9~{S},13~{S},14~{S},16~{E})-13-methyl-3-oxidanyl-16-(phenylmethylidene)-6,7,8,9,11,12,14,15-octahydrocyclopenta[ a]phenanthren-17-one, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-07-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
6D72
 
 | | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Yersinia pestis in complex with calcium ions. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, MALONATE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-23 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Yersinia pestis in complex with calcium ions.
To Be Published
|
|
6WGM
 
 | | Crystal structure of a marine metagenome TRAP solute binding protein specific for pyroglutamate (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, scf7180008839099) in complex with co-purified pyroglutamate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Fedorov, E, Vetting, M.W, Hogle, S.L, Dupont, C.L, Almo, S.C, Ghosh, A. | | Deposit date: | 2020-04-05 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, GOS_140), in complex with co-purified pyroglutamate
To Be Published
|
|
6WGX
 
 | | Cocrystal of BRD4(D1) with a selective inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-(1-{1-[2-(dimethylamino)ethyl]piperidin-4-yl}-4-[4-(trifluoromethyl)phenyl]-1H-imidazol-5-yl)-N-(3,5-dimethylphenyl)pyrimidin-2-amine, Bromodomain-containing protein 4 | | Authors: | Johnson, J.A, Cui, H, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2020-04-06 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Selective N-Terminal BET Bromodomain Inhibitors by Targeting Non-Conserved Residues and Structured Water Displacement*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5L1M
 
 | | CASKIN2 SAM domain tandem | | Descriptor: | Caskin-2 | | Authors: | Donaldson, L.W, Kwan, J.J, Saridakis, V. | | Deposit date: | 2016-07-29 | | Release date: | 2016-08-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | A new mode of SAM domain mediated oligomerization observed in the CASKIN2 neuronal scaffolding protein.
Cell Commun. Signal, 14, 2016
|
|
5L20
 
 | |
6WQZ
 
 | | Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery | | Descriptor: | Autophagy-related protein 9A, Lauryl Maltose Neopentyl Glycol | | Authors: | Guardia, C.M, Tan, X, Lian, T, Rana, M.S, Zhou, W, Christenson, E.T, Lowry, A.J, Faraldo-Gomez, J.D, Bonifacino, J.S, Jiang, J, Banerjee, A. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of Human ATG9A, the Only Transmembrane Protein of the Core Autophagy Machinery.
Cell Rep, 31, 2020
|
|
6WV7
 
 | | Human VKOR with Chlorophacinone | | Descriptor: | Chlorophacinone, Vitamin K epoxide reductase, termini restrained by green fluorescent protein | | Authors: | Liu, S, Sukumar, N, Li, W. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.483 Å) | | Cite: | Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation.
Science, 371, 2021
|
|
6WXW
 
 | |
6DGB
 
 | | Crystal structure of the C-terminal catalytic domain of IS1535 TnpA, an IS607-like serine recombinase | | Descriptor: | IS607 family transposase IS1535 | | Authors: | Chen, W.Y, Hancock, S.P, Cascio, D, Johnson, R.C. | | Deposit date: | 2018-05-17 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Multiple serine transposase dimers assemble the transposon-end synaptic complex during IS607-family transposition.
Elife, 7, 2018
|
|
5L6F
 
 | | Xylooligosaccharide oxidase from Myceliophthora thermophila C1 in complex with Xylobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rozeboom, H.J, Ferrari, A.R, Fraaije, M.W. | | Deposit date: | 2016-05-30 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a Xylooligosaccharide Oxidase from Myceliophthora thermophila C1.
J.Biol.Chem., 291, 2016
|
|
6WEP
 
 | | Crystal structure of TS-DHFR from Cryptosporidium hominis with Apo-TS site | | Descriptor: | Bifunctional dihydrofolate reductase-thymidylate synthase, METHOTREXATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Czyzyk, D.J, Ruiz, V.G, Kumar, V.P, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Targeting the TS dimer interface in bifunctional Cryptosporidium hominis TS-DHFR from parasitic protozoa: Virtual screening identifies novel TS allosteric inhibitor
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6DLL
 
 | | 2.2 Angstrom Resolution Crystal Structure of P-Hydroxybenzoate Hydroxylase from Pseudomonas putida in Complex with FAD. | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-01 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural comparison of p-hydroxybenzoate hydroxylase (PobA) from Pseudomonas putida with PobA from other Pseudomonas spp. and other monooxygenases.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
