9F9L
 
 | | Crystal structure of MUS81-EME1 bound by compound 16. | | Descriptor: | 2-[2-[4-(cyanomethyl)phenyl]phenyl]-5-oxidanyl-6-oxidanylidene-1H-pyrimidine-4-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
9AUK
 
 | | Structure of SARS-CoV-2 Mpro mutant (A173V) in complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
8Z38
 
 | | FK506 binding protein 1B (including FK506) | | Descriptor: | 6-CARBOXYPIPERIDINE, peptidylprolyl isomerase | | Authors: | Liu, X.H, Zhao, W.H. | | Deposit date: | 2024-04-14 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | High-resolution cocrystallized crystal structures of FK506 and FK506-binding proteins
To Be Published
|
|
9ASY
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a m-chlorobenzyl 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-({N-[({(2S)-1-[(3-chlorophenyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[({(2S)-1-[(3-chlorophenyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
8Y3U
 
 | |
7MNY
 
 | | Crystal Structure of Nup358/RanBP2 Ran-binding domain 3 in complex with Ran-GPPNHP | | Descriptor: | E3 SUMO-protein ligase RanBP2, GTP-binding nuclear protein Ran, MAGNESIUM ION, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
8ZH8
 
 | | Human GPR103 -Gq complex bound to QRFP26 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2,Guanine nucleotide-binding protein G(i) subunit alpha-2,Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Iwama, A, Akasaka, H, Sano, F.K, Oshima, H.S, Shihoya, W, Nureki, O. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structure and dynamics of the pyroglutamylated RF-amide peptide QRFP receptor GPR103.
Nat Commun, 15, 2024
|
|
8YY8
 
 | | Fzd7 -Gs complex | | Descriptor: | Frizzled-7, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Chen, B, Xu, L, Han, G.W, Xu, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Cryo-EM structure of constitutively active human Frizzled 7 in complex with heterotrimeric G s .
Cell Res., 31, 2021
|
|
9AUO
 
 | | Structure of SARS-CoV-2 Mpro mutant (L50F,T304I) | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9F98
 
 | | Crystal structure of MUS81-EME1, apo form. | | Descriptor: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81 | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
9ILD
 
 | | Complex structure of Cap3 and CD-NTase | | Descriptor: | Cyclic GMP-AMP synthase, Peptidase, ZINC ION | | Authors: | Li, F.Q, Ma, W.F. | | Deposit date: | 2024-06-29 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.179 Å) | | Cite: | Structural and functional characterization of the bacterial Cap3 enzyme in deconjugation and regulation of the cyclic dinucleotide transferase CD-NTase
To Be Published
|
|
8ZWV
 
 | | The Crystal Structure of carbonic anhydrase II from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Carbonic anhydrase 2, ZINC ION, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Bao, C. | | Deposit date: | 2024-06-13 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystal Structure of carbonic anhydrase II from Biortus.
To Be Published
|
|
9AUN
 
 | | Structure of SARS-CoV-2 Mpro mutant (T21I,T304I) | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Gajiwala, K.S, Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2024-02-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | In vitro selection and analysis of SARS-CoV-2 nirmatrelvir resistance mutations contributing to clinical virus resistance surveillance.
Sci Adv, 10, 2024
|
|
9CA1
 
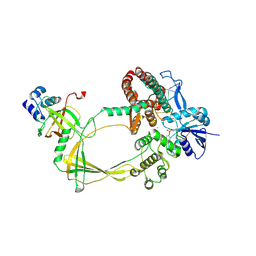 | | Human TOP3B-TDRD3 core complex in DNA religation state | | Descriptor: | DNA (5'-D(*AP*TP*T)-3'), DNA (5'-D(P*TP*AP*CP*TP*AP*AP*A)-3'), DNA topoisomerase 3-beta-1, ... | | Authors: | Yang, X, Chen, X, Yang, W, Pommier, Y. | | Deposit date: | 2024-06-16 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural Insights into human Topoisomerase 3-beta DNA and RNA catalytic cycles and topo-gate dynamics
To Be Published
|
|
8YUV
 
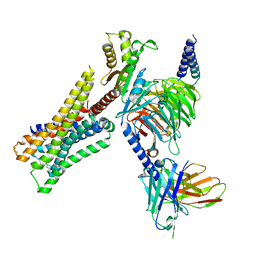 | | Cryo-EM structure of the immepip-bound H3R-Gi complex | | Descriptor: | 4-(1H-imidazol-5-ylmethyl)piperidine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shen, Q, Tang, X, Wen, X, Cheng, S, Xiao, P, Zang, S, Shen, D, Jiang, L, Zheng, Y, Zhang, H, Xu, H, Mao, C, Zhang, M, Hu, W, Sun, J, Chen, Z, Zhang, Y. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular Determinant Underlying Selective Coupling of Primary G-Protein by Class A GPCRs.
Adv Sci, 11, 2024
|
|
8XOT
 
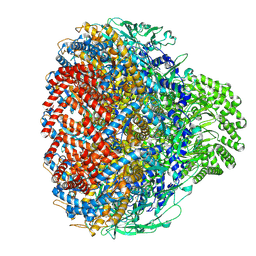 | | Prohead portal of bacteriophage lambda | | Descriptor: | Portal protein B | | Authors: | Wang, J.W, Gu, Z.W. | | Deposit date: | 2024-01-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural morphing in the viral portal vertex of bacteriophage lambda.
J.Virol., 98, 2024
|
|
8ENB
 
 | |
8YR3
 
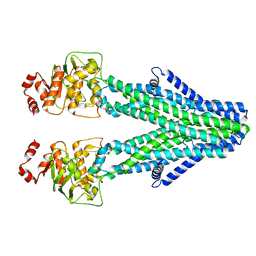 | | Cryo-EM structure of the human ABCB6 in complex with Cd(II):GSH | | Descriptor: | ATP-binding cassette sub-family B member 6, CADMIUM ION, GLUTATHIONE | | Authors: | Choi, S.H, Lee, S.S, Lee, H.Y, Kim, S, Kim, J.W, Jin, M.S. | | Deposit date: | 2024-03-20 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of cadmium-bound human ABCB6.
Commun Biol, 7, 2024
|
|
8YKW
 
 | | Cryo-EM structure of succinate receptor SUCR1 bound to succinic acid | | Descriptor: | Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, C, Liu, H, Li, J, Zhu, H, Fu, W, Xu, H.E. | | Deposit date: | 2024-03-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Molecular basis of ligand recognition and activation of the human succinate receptor SUCR1.
Cell Res., 34, 2024
|
|
9B3R
 
 | | The structure of human cardiac F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha cardiac muscle 1, ... | | Authors: | Doran, M.H, Sousa, D, Rynkiewicz, M.J, Lehman, W, Cammarato, A. | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of human cardiac actin
To Be Published
|
|
9ATJ
 
 | | Crystal structure of MERS 3CL protease in complex with a m-chlorobenzyl 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-({N-[({(2S)-1-[(3-chlorophenyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[({(2S)-1-[(3-chlorophenyl)methyl]-5-oxopyrrolidin-2-yl}methoxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|
9EOF
 
 | | Structure of the human INTS5/8/10/15 subcomplex | | Descriptor: | Integrator complex subunit 10, Integrator complex subunit 15, Integrator complex subunit 5, ... | | Authors: | Razew, M, Galej, W.P. | | Deposit date: | 2024-03-14 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural basis of the Integrator complex assembly and association with transcription factors.
Mol.Cell, 84, 2024
|
|
9EP4
 
 | | Structure of Integrator subcomplex INTS5/8/15 | | Descriptor: | Integrator complex subunit 15, Integrator complex subunit 5, Integrator complex subunit 8 | | Authors: | Razew, M, Galej, W.P. | | Deposit date: | 2024-03-17 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of the Integrator complex assembly and association with transcription factors.
Mol.Cell, 84, 2024
|
|
8XB4
 
 | |
8YM1
 
 | | Structure of SADS-CoV Virus Nucleocapsid Protein | | Descriptor: | nucleocapsid phosphoprotein | | Authors: | Zhang, Y, Wu, F, Xu, W. | | Deposit date: | 2024-03-08 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unraveling the assembly mechanism of SADS-CoV virus nucleocapsid protein: insights from RNA binding, dimerization, and epitope diversity profiling.
J.Virol., 2024
|
|
