5BMG
 
 | | Nitroxide Spin Labels in Protein GB1: E15 Mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Immunoglobulin G-binding protein G, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Cunningham, T.C, Horne, W.S, Saxena, S. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rotameric preferences of a protein spin label at edge-strand beta-sheet sites.
Protein Sci., 25, 2016
|
|
3DMK
 
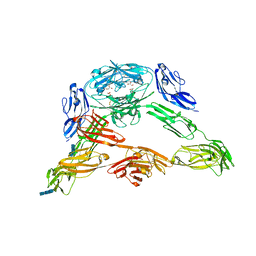 | | Crystal structure of Down Syndrome Cell Adhesion Molecule (DSCAM) isoform 1.30.30, N-terminal eight Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down Syndrome Cell Adhesion Molecule (DSCAM) isoform 1.30.30, ... | | Authors: | Sawaya, M.R, Wojtowicz, W.M, Eisenberg, D, Zipursky, S.L. | | Deposit date: | 2008-07-01 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.19 Å) | | Cite: | A double S shape provides the structural basis for the extraordinary binding specificity of Dscam isoforms.
Cell(Cambridge,Mass.), 134, 2008
|
|
5GAI
 
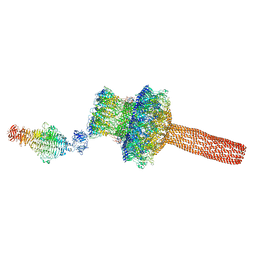 | | Probabilistic Structural Models of Mature P22 Bacteriophage Portal, Hub, and Tailspike proteins | | Descriptor: | Peptidoglycan hydrolase gp4, Portal protein, Tail fiber protein | | Authors: | Pintilie, G, Chen, D.H, Haase-Pettingell, C.A, King, J.A, Chiu, W. | | Deposit date: | 2015-12-01 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10.5 Å) | | Cite: | Resolution and Probabilistic Models of Components in CryoEM Maps of Mature P22 Bacteriophage.
Biophys.J., 110, 2016
|
|
1ZID
 
 | |
5GTP
 
 | |
5ZHZ
 
 | | Crystal structure of the apurinic/apyrimidinic endonuclease IV from Mycobacterium tuberculosis | | Descriptor: | Probable endonuclease 4, SULFATE ION, ZINC ION | | Authors: | Zhang, W, Xu, Y, Yan, M, Li, S, Wang, H, Yang, H, Zhou, W, Rao, Z. | | Deposit date: | 2018-03-13 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystal structure of the apurinic/apyrimidinic endonuclease IV from Mycobacterium tuberculosis.
Biochem. Biophys. Res. Commun., 498, 2018
|
|
3OFT
 
 | | Crystal Structure of Cytochrome P450 CYP101C1 | | Descriptor: | (2R,5R)-hexane-2,5-diol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhou, W, Ma, M, Bell, S.G, Yang, W, Hao, Y, Rees, N.H, Bartlam, M, Wong, L.-L, Rao, Z. | | Deposit date: | 2010-08-16 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of CYP101C1 from Novosphingobium aromaticivorans DSM12444.
Chembiochem, 12, 2011
|
|
1L89
 
 | |
4RXZ
 
 | |
8K6Z
 
 | | NMR structure of human leptin | | Descriptor: | Leptin | | Authors: | Fan, X, Qin, R, Yuan, W, Fan, J, Huang, W, Lin, Z. | | Deposit date: | 2023-07-26 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human leptin reveals a conformational plasticity important for receptor recognition.
Structure, 32, 2024
|
|
2AI6
 
 | |
3TU8
 
 | | Crystal Structure of the Burkholderia Lethal Factor 1 (BLF1) | | Descriptor: | BROMIDE ION, Burkholderia Lethal Factor 1 (BLF1) | | Authors: | Cruz, A, Hautbergue, G.M, Artymiuk, P.J, Baker, P.J, Chang, C.T, Mahadi, N.M, Mobbs, G.W, Mohamed, R, Nathan, S, Partridge, L.J, Raih, M.F, Ruzheinikov, S.N, Sedelnikova, S.E, Wilson, S.A, Rice, D.W. | | Deposit date: | 2011-09-16 | | Release date: | 2011-11-30 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | A Burkholderia pseudomallei toxin inhibits helicase activity of translation factor eIF4A.
Science, 334, 2011
|
|
5DAJ
 
 | | Crystal structure of NalD, the secondary repressor of MexAB-OprM multidrug efflux pump in Pseudomonas aeruginosa | | Descriptor: | NalD | | Authors: | Chen, W.Z, Wang, D, Huang, S.Q, Hu, Q.Y, Liu, X.C, Gan, J.H, Chen, H. | | Deposit date: | 2015-08-20 | | Release date: | 2016-04-20 | | Last modified: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Novobiocin binding to NalD induces the expression of the MexAB-OprM pump in Pseudomonas aeruginosa
Mol.Microbiol., 100, 2016
|
|
4RM4
 
 | | The crystal structure of the versatile cytochrome P450 enzyme CYP109B1 from Bacillus subtilis | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhouw, W.H, Zhang, A.L, Zhang, T, Hall, E.A, Hutchinson, S, Cryle, M.J, Wong, L.-L, Bell, S.G. | | Deposit date: | 2014-10-18 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | The crystal structure of the versatile cytochrome P450 enzyme CYP109B1 from Bacillus subtilis.
Mol Biosyst, 11, 2015
|
|
4NTJ
 
 | | Structure of the human P2Y12 receptor in complex with an antithrombotic drug | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, P2Y purinoceptor 12,Soluble cytochrome b562,P2Y purinoceptor 12, ... | | Authors: | Zhang, K, Zhang, J, Gao, Z.-G, Zhang, D, Zhu, L, Han, G.W, Moss, S.M, Paoletta, S, Kiselev, E, Lu, W, Fenalti, G, Zhang, W, Muller, C.E, Yang, H, Jiang, H, Cherezov, V, Katritch, V, Jacobson, K.A, Stevens, R.C, Wu, B, Zhao, Q, GPCR Network (GPCR) | | Deposit date: | 2013-12-02 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure of the human P2Y12 receptor in complex with an antithrombotic drug
Nature, 509, 2014
|
|
2ARK
 
 | | Structure of a flavodoxin from Aquifex aeolicus | | Descriptor: | Flavodoxin, GLYCEROL, PHOSPHATE ION | | Authors: | Cuff, M.E, Quartey, P, Zhou, M, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-19 | | Release date: | 2005-10-25 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a flavodoxin from Aquifex aeolicus
To be Published
|
|
5CSM
 
 | | YEAST CHORISMATE MUTASE, T226S MUTANT, COMPLEX WITH TRP | | Descriptor: | CHORISMATE MUTASE, TRYPTOPHAN | | Authors: | Straeter, N, Schnappauf, G, Braus, G, Lipscomb, W.N. | | Deposit date: | 1997-07-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanisms of catalysis and allosteric regulation of yeast chorismate mutase from crystal structures.
Structure, 5, 1997
|
|
1ETR
 
 | | REFINED 2.3 ANGSTROMS X-RAY CRYSTAL STRUCTURE OF BOVINE THROMBIN COMPLEXES FORMED WITH THE BENZAMIDINE AND ARGININE-BASED THROMBIN INHIBITORS NAPAP, 4-TAPAP AND MQPA: A STARTING POINT FOR IMPROVING ANTITHROMBOTICS | | Descriptor: | EPSILON-THROMBIN, amino{[(4S)-5-[(2R,4R)-2-carboxy-4-methylpiperidin-1-yl]-4-({[(3R)-3-methyl-1,2,3,4-tetrahydroquinolin-8-yl]sulfonyl}amino)-5-oxopentyl]amino}methaniminium | | Authors: | Bode, W, Brandstetter, H. | | Deposit date: | 1992-07-06 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined 2.3 A X-ray crystal structure of bovine thrombin complexes formed with the benzamidine and arginine-based thrombin inhibitors NAPAP, 4-TAPAP and MQPA. A starting point for improving antithrombotics.
J.Mol.Biol., 226, 1992
|
|
2B7Y
 
 | | Fava Bean Lectin-Glucose Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Favin alpha chain, ... | | Authors: | Reeke Jr, G.N, Becker, J.W. | | Deposit date: | 2005-10-05 | | Release date: | 2005-10-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-dimensional structure of favin: saccharide binding-cyclic permutation in leguminous lectins
Science, 234, 1986
|
|
3H8E
 
 | |
2CJ1
 
 | | chloroperoxidase complexed with formate (ethylene glycol cryoprotectant) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuhnel, K, Blankenfeldt, W, Terner, J, Schlichting, I. | | Deposit date: | 2006-03-26 | | Release date: | 2006-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Chloroperoxidase with its Bound Substrates and Complexed with Formate, Acetate, and Nitrate.
J.Biol.Chem., 281, 2006
|
|
1ZK3
 
 | | Triclinic crystal structure of the apo-form of R-specific alcohol dehydrogenase (mutant G37D) from Lactobacillus brevis | | Descriptor: | MAGNESIUM ION, R-specific alcohol dehydrogenase | | Authors: | Schlieben, N.H, Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Atomic Resolution Structures of R-specific Alcohol Dehydrogenase from Lactobacillus brevis Provide the Structural Bases of its Substrate and Cosubstrate Specificity
J.Mol.Biol., 349, 2005
|
|
5VVV
 
 | | Structural Investigations of the Substrate Specificity of Human O-GlcNAcase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein O-GlcNAcase, a-crystallin B | | Authors: | Li, B, Jiang, J, Li, H, Hu, C.-W. | | Deposit date: | 2017-05-20 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the substrate binding adaptability and specificity of human O-GlcNAcase.
Nat Commun, 8, 2017
|
|
5CLO
 
 | |
5GTB
 
 | |
