5NGE
 
 | | Crystal structure of USP7 in complex with the non-covalent inhibitor, FT671 | | Descriptor: | 5-[[1-[(3~{S})-4,4-bis(fluoranyl)-3-(3-fluoranylpyrazol-1-yl)butanoyl]-4-oxidanyl-piperidin-4-yl]methyl]-1-(4-fluorophenyl)pyrazolo[3,4-d]pyrimidin-4-one, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Turnbull, A.P, Krajewski, W.W, Ioannidis, S, Kessler, B.M, Komander, D. | | Deposit date: | 2017-03-17 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular basis of USP7 inhibition by selective small-molecule inhibitors.
Nature, 550, 2017
|
|
5NIL
 
 | | Structure of the MacAB-TolC ABC-type tripartite multidrug efflux pump-MacB section | | Descriptor: | Macrolide export ATP-binding/permease protein MacB, Macrolide export protein MacA, Outer membrane protein TolC | | Authors: | Fitzpatrick, A.W.P, Llabres, S, Neuberger, A, Blaza, J.N, Bai, X.-C, Okada, U, Murakami, S, van Veen, H.W, Zachariae, U, Scheres, S.H.W, Luisi, B.F, Du, D. | | Deposit date: | 2017-03-24 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Structure of the MacAB-TolC ABC-type tripartite multidrug efflux pump.
Nat Microbiol, 2, 2017
|
|
6TBF
 
 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | (1~{S},2~{S},3~{S},4~{R},5~{R})-4-(hydroxymethyl)-5-(octylamino)cyclopentane-1,2,3-triol, ACETATE ION, Beta-galactosidase, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2019-11-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic Insights into the Chaperoning of Human Lysosomal-Galactosidase Activity: Highly Functionalized Aminocyclopentanes and C -5a-Substituted Derivatives of 4- epi -Isofagomine.
Molecules, 25, 2020
|
|
1TRP
 
 | |
5VC8
 
 | | Crystal structure of the WHSC1 PWWP1 domain | | Descriptor: | DNA (5'-D(P*CP*TP*(DN))-3'), Histone-lysine N-methyltransferase NSD2, UNKNOWN ATOM OR ION, ... | | Authors: | Qin, S, Tempel, W, Dong, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-28 | | Last modified: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Histone and DNA binding ability studies of the NSD subfamily of PWWP domains.
Biochem.Biophys.Res.Commun., 569, 2021
|
|
6TBI
 
 | | Structure of a beta galactosidase with inhibitor | | Descriptor: | (1S,2S,3S,6R)-4-(hydroxymethyl)-6-(octylamino)cyclohex-4-ene-1,2,3-triol, ACETATE ION, Beta-galactosidase, ... | | Authors: | Offen, W, Davies, G. | | Deposit date: | 2019-11-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Mechanistic Insights into the Chaperoning of Human Lysosomal-Galactosidase Activity: Highly Functionalized Aminocyclopentanes and C -5a-Substituted Derivatives of 4- epi -Isofagomine.
Molecules, 25, 2020
|
|
1TV6
 
 | | HIV-1 Reverse Transcriptase Complexed with CP-94,707 | | Descriptor: | 3-[4-(2-METHYL-IMIDAZO[4,5-C]PYRIDIN-1-YL)BENZYL]-3H-BENZOTHIAZOL-2-ONE, reverse transcriptase p51 subunit, reverse transcriptase p66 subunit | | Authors: | Pata, J.D, Stirtan, W.G, Goldstein, S.W, Steitz, T.A. | | Deposit date: | 2004-06-28 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of HIV-1 reverse transcriptase bound to an inhibitor active against mutant RTs resistant to other non-nucleoside inhibitors
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
4ZWJ
 
 | | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser | | Descriptor: | Chimera protein of human Rhodopsin, mouse S-arrestin, and T4 Endolysin | | Authors: | Kang, Y, Zhou, X.E, Gao, X, He, Y, Liu, W, Ishchenko, A, Barty, A, White, T.A, Yefanov, O, Han, G.W, Xu, Q, de Waal, P.W, Ke, J, Tan, M.H.E, Zhang, C, Moeller, A, West, G.M, Pascal, B, Eps, N.V, Caro, L.N, Vishnivetskiy, S.A, Lee, R.J, Suino-Powell, K.M, Gu, X, Pal, K, Ma, J, Zhi, X, Boutet, S, Williams, G.J, Messerschmidt, M, Gati, C, Zatsepin, N.A, Wang, D, James, D, Basu, S, Roy-Chowdhury, S, Conrad, C, Coe, J, Liu, H, Lisova, S, Kupitz, C, Grotjohann, I, Fromme, R, Jiang, Y, Tan, M, Yang, H, Li, J, Wang, M, Zheng, Z, Li, D, Howe, N, Zhao, Y, Standfuss, J, Diederichs, K, Dong, Y, Potter, C.S, Carragher, B, Caffrey, M, Jiang, H, Chapman, H.N, Spence, J.C.H, Fromme, P, Weierstall, U, Ernst, O.P, Katritch, V, Gurevich, V.V, Griffin, P.R, Hubbell, W.L, Stevens, R.C, Cherezov, V, Melcher, K, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser.
Nature, 523, 2015
|
|
2AT1
 
 | |
2FEQ
 
 | | orally active thrombin inhibitors | | Descriptor: | Decapeptide Hirudin Analogue, N-(CARBOXYMETHYL)-3-CYCLOHEXYL-D-ALANYL-N-({4-[(E)-AMINO(IMINO)METHYL]-1,3-THIAZOL-2-YL}METHYL)-L-PROLINAMIDE, Thrombin heavy chain, ... | | Authors: | Mack, H, Baucke, D, Hornberger, W, Lange, U.E.W, Hoeffken, H.W. | | Deposit date: | 2005-12-16 | | Release date: | 2006-08-08 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Orally active thrombin inhibitors. Part 1: optimization of the P1-moiety
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1WQ1
 
 | | RAS-RASGAP COMPLEX | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, H-RAS, ... | | Authors: | Scheffzek, K, Ahmadian, M.R, Kabsch, W, Wiesmueller, L, Lautwein, A, Schmitz, F, Wittinghofer, A. | | Deposit date: | 1997-07-03 | | Release date: | 1998-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants.
Science, 277, 1997
|
|
2LZM
 
 | |
1CA6
 
 | | INTERCALATION SITE OF HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SSO7D/SAC7D BOUND TO DNA | | Descriptor: | 5'-D(*GP*TP*GP*AP*TP*CP*GP*C)-3', CHROMOSOMAL PROTEIN SAC7D | | Authors: | Su, S, Gao, Y.-G, Robinson, H, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 1999-02-23 | | Release date: | 2000-02-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the chromosomal proteins Sso7d/Sac7d bound to DNA containing T-G mismatched base-pairs
J.Mol.Biol., 303, 2000
|
|
6NMY
 
 | | A Cytokine-receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit beta, ... | | Authors: | Dhagat, U, Kan, W.L, Hercus, T.R, Broughton, S.E, Nero, T.L, Lopez, A.F, Parker, M.W. | | Deposit date: | 2019-01-13 | | Release date: | 2020-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | Distinct Assemblies of Heterodimeric Cytokine Receptors Govern Stemness Programs in Leukemia.
Cancer Discov, 13, 2023
|
|
6QQP
 
 | | Aplysia californica AChBP in complex with 2-Fluoro-(carbamoylpyridinyl)deschloroepibatidine analogue (2) | | Descriptor: | 1,2-ETHANEDIOL, 2-[5-[(1~{R},2~{R},4~{S})-7-azabicyclo[2.2.1]heptan-2-yl]-2-fluoranyl-pyridin-3-yl]pyridine-4-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bueno, R.V, Davis, S, Dawson, A, Hunter, W.N. | | Deposit date: | 2019-02-18 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Interactions between 2'-fluoro-(carbamoylpyridinyl)deschloroepibatidine analogues and acetylcholine-binding protein inform on potent antagonist activity against nicotinic receptors
Acta Crystallogr.,Sect.D, 78, 2022
|
|
5IMI
 
 | | Crystal structure of S303A Aspergillus terreus aristolochene synthase complexed with (1S,8S,9aR)-1,9a-dimethyl-8-(prop-1-en-2-yl)decahydroquinolizin-5-ium | | Descriptor: | (1S,5S,8S,9aR)-1,9a-dimethyl-8-(prop-1-en-2-yl)octahydro-2H-quinolizinium, Aristolochene synthase, GLYCEROL, ... | | Authors: | Chen, M, Christianson, D.W. | | Deposit date: | 2016-03-06 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.462 Å) | | Cite: | Probing the Role of Active Site Water in the Sesquiterpene Cyclization Reaction Catalyzed by Aristolochene Synthase.
Biochemistry, 55, 2016
|
|
3EBX
 
 | | REFINEMENT AT 1.4 ANGSTROMS RESOLUTION OF A MODEL OF ERABUTOXIN B. TREATMENT OF ORDERED SOLVENT AND DISCRETE DISORDER | | Descriptor: | ERABUTOXIN B, SULFATE ION | | Authors: | Smith, J.L, Corfield, P.W.R, Hendrickson, W.A, Low, B.W. | | Deposit date: | 1988-01-15 | | Release date: | 1988-04-16 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Refinement at 1.4 A resolution of a model of erabutoxin b: treatment of ordered solvent and discrete disorder.
Acta Crystallogr.,Sect.A, 44, 1988
|
|
1BZY
 
 | | HUMAN HGPRTASE WITH TRANSITION STATE INHIBITOR | | Descriptor: | HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, MAGNESIUM ION, PHOSPHORIC ACID MONO-[5-(2-AMINO-4-OXO-4,5-DIHYDRO-3H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-3,4-DIHYDROXY-PYRROLIDIN-2-YLMETHYL] ESTER, ... | | Authors: | Shi, W, Li, C, Tyler, P.C, Furneaux, R.H, Grubmeyer, C, Schramm, V.L, Almo, S.C. | | Deposit date: | 1998-11-05 | | Release date: | 1999-06-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A structure of human hypoxanthine-guanine phosphoribosyltransferase in complex with a transition-state analog inhibitor.
Nat.Struct.Biol., 6, 1999
|
|
1T8A
 
 | | USE OF SEQUENCE DUPLICATION TO ENGINEER A LIGAND-TRIGGERED LONG-DISTANCE MOLECULAR SWITCH IN T4 Lysozyme | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, GUANIDINE, ... | | Authors: | Yousef, M.S, Baase, W.A, Matthews, B.W. | | Deposit date: | 2004-05-11 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Use of sequence duplication to engineer a ligand-triggered, long-distance molecular switch in T4 lysozyme.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1CGX
 
 | |
1CVD
 
 | |
1CET
 
 | | CHLOROQUINE BINDS IN THE COFACTOR BINDING SITE OF PLASMODIUM FALCIPARUM LACTATE DEHYDROGENASE. | | Descriptor: | N4-(7-CHLORO-QUINOLIN-4-YL)-N1,N1-DIETHYL-PENTANE-1,4-DIAMINE, PROTEIN (L-LACTATE DEHYDROGENASE) | | Authors: | Read, J.A, Wilkinson, K.W, Tranter, R, Sessions, R.B, Brady, R.L. | | Deposit date: | 1999-03-10 | | Release date: | 1999-03-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Chloroquine binds in the cofactor binding site of Plasmodium falciparum lactate dehydrogenase.
J.Biol.Chem., 274, 1999
|
|
8P6I
 
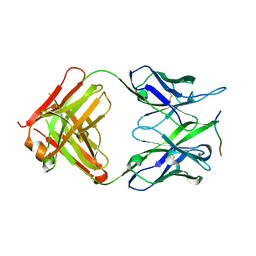 | | Crystal structure of the 139H2 Fab fragment bound to Muc1 peptide epitope | | Descriptor: | 139H2 HC, 139H2 LC, Mucin-1 | | Authors: | Beugelink, J.W, Peng, W, Siborova, M, Pronker, M.F, Snijder, J, Janssen, B.J.C. | | Deposit date: | 2023-05-26 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Reverse-engineering the anti-MUC1 antibody 139H2 by mass spectrometry-based de novo sequencing.
Life Sci Alliance, 7, 2024
|
|
3IQC
 
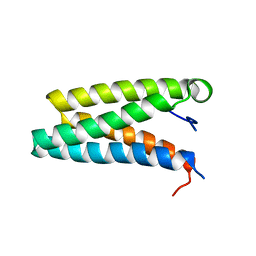 | |
1CVH
 
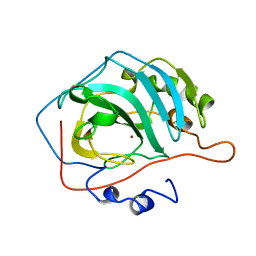 | |
