1SHW
 
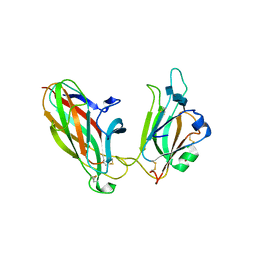 | | EphB2 / EphrinA5 Complex Structure | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin type-B receptor 2, Ephrin-A5, ... | | 著者 | Himanen, J.P, Chumley, M.J, Lackmann, M, Li, C, Barton, W.A, Jeffrey, P.D, Vearing, C, Geleick, D, Feldheim, D.A, Boyd, A.W. | | 登録日 | 2004-02-26 | | 公開日 | 2004-05-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Repelling class discrimination: ephrin-A5 binds to and activates EphB2 receptor signaling
Nat.Neurosci., 7, 2004
|
|
8X5L
 
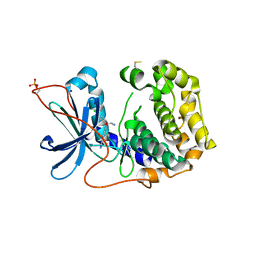 | | The Crystal Structure of PRKACA from Biortus. | | 分子名称: | (2S)-2-(4-chlorophenyl)-2-hydroxy-2-[4-(1H-pyrazol-4-yl)phenyl]ethanaminium, SODIUM ION, cAMP-dependent protein kinase catalytic subunit alpha | | 著者 | Wang, F, Cheng, W, Lv, Z, Lin, D, Pan, W. | | 登録日 | 2023-11-17 | | 公開日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | The Crystal Structure of PRKACA from Biortus.
To Be Published
|
|
7F8T
 
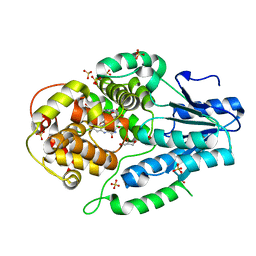 | | Re-refinement of the 2XRY X-ray structure of archaeal class II CPD photolyase from Methanosarcina mazei | | 分子名称: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | 著者 | Maestre-Reyna, M, Yang, C.-H, Huang, W.C, Nango, E, Gusti-Ngurah-Putu, E.-P, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Sugahara, M, Owada, S, Joti, Y, Tanaka, R, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | 登録日 | 2021-07-02 | | 公開日 | 2022-03-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
1G7Y
 
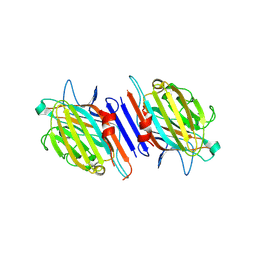 | | THE CRYSTAL STRUCTURE OF THE 58KD VEGETATIVE LECTIN FROM THE TROPICAL LEGUME DOLICHOS BIFLORUS | | 分子名称: | CALCIUM ION, MANGANESE (II) ION, STEM/LEAF LECTIN DB58, ... | | 著者 | Buts, L, Hamelryck, T.W, Loris, R, Dao-Thi, M.-H, Wyns, L, Etzler, M.E. | | 登録日 | 2000-11-15 | | 公開日 | 2000-11-29 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Weak protein-protein interactions in lectins: the crystal structure of a vegetative lectin from the legume Dolichos biflorus.
J.Mol.Biol., 309, 2001
|
|
6FKX
 
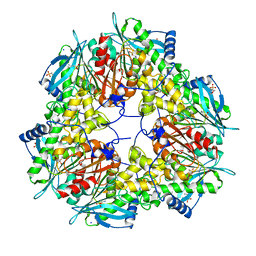 | | Crystal structure of an acetyl xylan esterase from a desert metagenome | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acetyl xylan esterase, FORMIC ACID, ... | | 著者 | Adesioye, F.A, Makhalanyane, T.P, Vikram, S, Sewell, B.T, Schubert, W, Cowan, D.A. | | 登録日 | 2018-01-24 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Structural Characterization and Directed Evolution of a Novel Acetyl Xylan Esterase Reveals Thermostability Determinants of the Carbohydrate Esterase 7 Family.
Appl. Environ. Microbiol., 84, 2018
|
|
1FXV
 
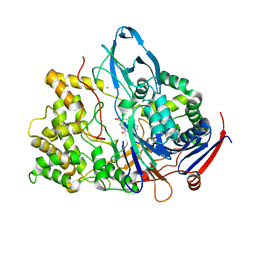 | | PENICILLIN ACYLASE MUTANT IMPAIRED IN CATALYSIS WITH PENICILLIN G IN THE ACTIVE SITE | | 分子名称: | CALCIUM ION, PENICILLIN ACYLASE, PENICILLIN G | | 著者 | Alkema, W.B, Hensgens, C.M, Kroezinga, E.H, de Vries, E, Floris, R, van der Laan, J.M, Dijkstra, B.W, Janssen, D.B. | | 登録日 | 2000-09-27 | | 公開日 | 2001-03-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and site-directed mutagenesis studies.
Protein Eng., 13, 2000
|
|
6V3B
 
 | |
3KSV
 
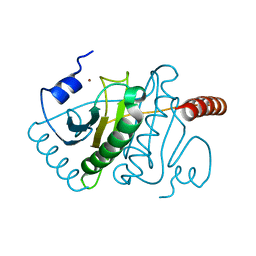 | |
1G1W
 
 | | T4 LYSOZYME MUTANT C54T/C97A/Q105M | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | 著者 | Quillin, M.L, Matthews, B.W. | | 登録日 | 2000-10-13 | | 公開日 | 2001-05-09 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
1S1S
 
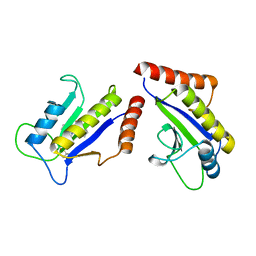 | | Crystal Structure of ZipA in complex with indoloquinolizin 10b | | 分子名称: | Cell division protein zipA, N-{3-[(12bS)-7-oxo-1,3,4,6,7,12b-hexahydroindolo[2,3-a]quinolizin-12(2H)-yl]propyl}propane-2-sulfonamide | | 著者 | Jennings, L.D, Foreman, K.W, Rush III, T.S, Tsao, D.H, Mosyak, L, Li, Y, Sukhdeo, M.N, Ding, W, Dushin, E.G, Kenny, C.H, Moghazeh, S.L, Petersen, P.J, Ruzin, A.V, Tuckman, M, Sutherland, A.G. | | 登録日 | 2004-01-07 | | 公開日 | 2005-01-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Design and synthesis of indolo[2,3-a]quinolizin-7-one inhibitors of the ZipA-FtsZ interaction
BIOORG.MED.CHEM.LETT., 14, 2004
|
|
7FIV
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidA and CidBND1-ND2 from wPip(Tunis) | | 分子名称: | CidA_I gamma/2 protein, CidB_I b/2 protein | | 著者 | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-08-01 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIW
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidAwMel(ST) and CidBND1-ND2 from wPip(Pel) | | 分子名称: | ULP_PROTEASE domain-containing protein, bacteria factor 4,CidA I(Zeta/1) protein | | 著者 | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-08-01 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIU
 
 | | Crystal structure of the DUB domain of Wolbachia cytoplasmic incompatibility factor CidB from wMel | | 分子名称: | ULP_PROTEASE domain-containing protein | | 著者 | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-08-01 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIT
 
 | | Crystal structure of Wolbachia cytoplasmic incompatibility factor CidA from wMel | | 分子名称: | bacteria factor 1 | | 著者 | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-08-01 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7AE1
 
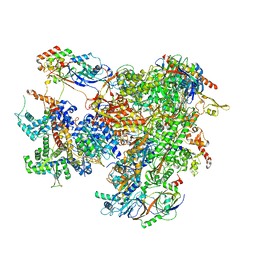 | | Cryo-EM structure of human RNA Polymerase III elongation complex 1 | | 分子名称: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | 著者 | Girbig, M, Misiaszek, A.D, Vorlaender, M.K, Mueller, C.W. | | 登録日 | 2020-09-17 | | 公開日 | 2021-02-03 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3JBT
 
 | | Atomic structure of the Apaf-1 apoptosome | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apoptotic protease-activating factor 1, Cytochrome c, ... | | 著者 | Zhou, M, Li, Y, Hu, Q, Bai, X, Huang, W, Yan, C, Scheres, S.H.W, Shi, Y. | | 登録日 | 2015-10-15 | | 公開日 | 2015-11-18 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Atomic structure of the apoptosome: mechanism of cytochrome c- and dATP-mediated activation of Apaf-1
Genes Dev., 29, 2015
|
|
6MPV
 
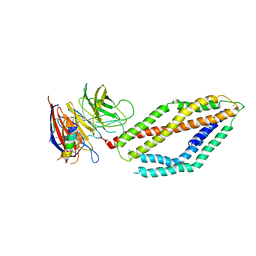 | |
6FPS
 
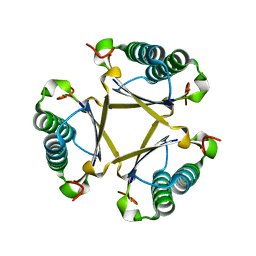 | |
1G06
 
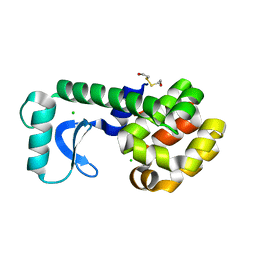 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT V149S | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | 著者 | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | 登録日 | 2000-10-05 | | 公開日 | 2001-05-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
1G0L
 
 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT T152V | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | 著者 | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | 登録日 | 2000-10-06 | | 公開日 | 2001-05-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
6FNU
 
 | | Structure of S. cerevisiae Methylenetetrahydrofolate reductase 1, catalytic domain | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Methylenetetrahydrofolate reductase 1 | | 著者 | Kopec, J, Rembeza, E, Bezerra, G.A, Newman, J, Bountra, C, Froese, D.S, Baumgartner, M, Yue, W.W, Structural Genomics Consortium (SGC) | | 登録日 | 2018-02-05 | | 公開日 | 2018-03-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Structural basis for the regulation of human 5,10-methylenetetrahydrofolate reductase by phosphorylation and S-adenosylmethionine inhibition.
Nat Commun, 9, 2018
|
|
4ZXU
 
 | | 2.85 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) H448F/P449M double mutant from Staphylococcus aureus in complex with NAD+ and BME-free Cys289 | | 分子名称: | Betaine-aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | 著者 | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-05-20 | | 公開日 | 2015-06-17 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | 2.85 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) H448F/P449M double mutant from Staphylococcus aureus in complex with NAD+ and BME-free Cys289.
To be Published
|
|
6MUV
 
 | | The structure of the Plasmodium falciparum 20S proteasome in complex with two PA28 activators | | 分子名称: | 20S proteasome alpha-1 subunit, 20S proteasome alpha-2 subunit, 20S proteasome alpha-3 subunit, ... | | 著者 | Metcalfe, R.D, Xie, S.C, Hanssen, E, Gillett, D.L, Leis, A.P, Tilley, L, Griffin, M.D.W. | | 登録日 | 2018-10-23 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | The structure of the PA28-20S proteasome complex from Plasmodium falciparum and implications for proteostasis.
Nat Microbiol, 4, 2019
|
|
1XH7
 
 | | Crystal Structures of Protein Kinase B Selective Inhibitors in Complex with Protein Kinase A and Mutants | | 分子名称: | N-[4-({4-[5-(3,3-DIMETHYLPIPERIDIN-1-YL)-2-HYDROXYBENZOYL]BENZOYL}AMINO)AZEPAN-3-YL]ISONICOTINAMIDE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | 著者 | Breitenlechner, C.B, Friebe, W.-G, Brunet, E, Werner, G, Graul, K, Thomas, U, Kuenkele, K.-P, Schaefer, W, Gassel, M, Bossemeyer, D, Huber, R, Engh, R.A, Masjost, B. | | 登録日 | 2004-09-17 | | 公開日 | 2005-09-17 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Design and crystal structures of protein kinase B-selective inhibitors in complex with protein kinase A and mutants
J.Med.Chem., 48, 2005
|
|
1G07
 
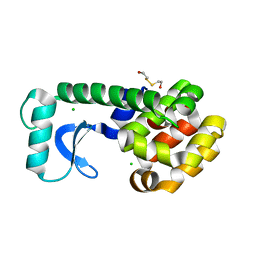 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT V149C | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | 著者 | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | 登録日 | 2000-10-05 | | 公開日 | 2001-05-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
