3U78
 
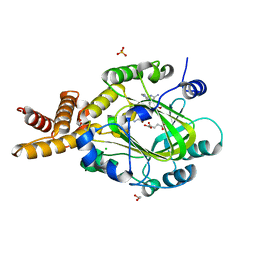 | | E67-2 selectively inhibits KIAA1718, a human histone H3 lysine 9 Jumonji demethylase | | 分子名称: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, 7-[(5-aminopentyl)oxy]-N~4~-(1-benzylpiperidin-4-yl)-N~2~-[3-(dimethylamino)propyl]-6-methoxyquinazoline-2,4-diamine, ... | | 著者 | Upadhyay, A.K, Cheng, X. | | 登録日 | 2011-10-13 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.689 Å) | | 主引用文献 | An Analog of BIX-01294 Selectively Inhibits a Family of Histone H3 Lysine 9 Jumonji Demethylases.
J.Mol.Biol., 416, 2012
|
|
2JVG
 
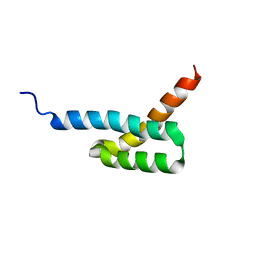 | | Structure of C3-binding domain 4 of Staphylococcus aureus protein Sbi | | 分子名称: | IgG-binding protein SBI | | 著者 | Upadhyay, A, Burman, J, Clark, E.A, van den Elsen, J.M.H, Bagby, S. | | 登録日 | 2007-09-20 | | 公開日 | 2008-06-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-function analysis of the C3 binding region of Staphylococcus aureus immune subversion protein Sbi.
J.Biol.Chem., 283, 2008
|
|
2JVH
 
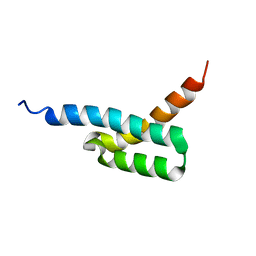 | | Structure of C3-binding domain 4 of S. aureus protein Sbi | | 分子名称: | IgG-binding protein SBI | | 著者 | Upadhyay, A, Burman, J, Clark, E.A, van den Elsen, J.M.H, Bagby, S. | | 登録日 | 2007-09-20 | | 公開日 | 2008-06-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-function analysis of the C3 binding region of Staphylococcus aureus immune subversion protein Sbi.
J.Biol.Chem., 283, 2008
|
|
2WY7
 
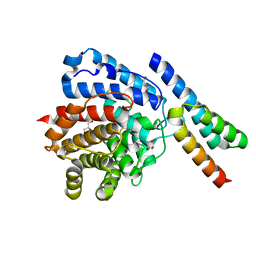 | | Staphylococcus aureus complement subversion protein Sbi-IV in complex with complement fragment C3d revealing an alternative binding mode | | 分子名称: | COMPLEMENT C3D FRAGMENT, GLYCEROL, IGG-BINDING PROTEIN | | 著者 | Clark, E.A, Crennell, S, Upadhyay, A, Mackay, J.D, Bagby, S, van den Elsen, J.M. | | 登録日 | 2009-11-13 | | 公開日 | 2010-12-01 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A Structural Basis for Staphylococcal Complement Subversion: X-Ray Structure of the Complement- Binding Domain of Staphylococcus Aureus Protein Sbi in Complex with Ligand C3D.
Mol.Immunol., 48, 2011
|
|
2WY8
 
 | | Staphylococcus aureus complement subversion protein Sbi-IV in complex with complement fragment C3d | | 分子名称: | COMPLEMENT C3D FRAGMENT, GLYCEROL, IGG-BINDING PROTEIN | | 著者 | Clark, E.A, Crennell, S, Upadhyay, A, Mackay, J.D, Bagby, S, van den Elsen, J.M. | | 登録日 | 2009-11-13 | | 公開日 | 2010-12-01 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | A Structural Basis for Staphylococcal Complement Subversion: X-Ray Structure of the Complement- Binding Domain of Staphylococcus Aureus Protein Sbi in Complex with Ligand C3D.
Mol.Immunol., 48, 2011
|
|
5TFR
 
 | | Crystal structure of Zika Virus NS5 protein | | 分子名称: | Genome polyprotein, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | 著者 | Longenecker, K.L, Upadhyay, A.K. | | 登録日 | 2016-09-26 | | 公開日 | 2016-10-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Crystal structure of full-length Zika virus NS5 protein reveals a conformation similar to Japanese encephalitis virus NS5.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
2L5T
 
 | |
8V15
 
 | | Human SIRT3 bound to p53-AMC peptide, Carba-NAD, and Honokiol | | 分子名称: | (1P)-3',5-di(prop-2-en-1-yl)[1,1'-biphenyl]-2,4'-diol, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, GLN-PRO-LYS-FDL, ... | | 著者 | Chakrabarti, R, Ghosh, A, Guan, X, Upadhyay, A, Dumpati, R.K, Munshi, S, Roy, S, Chall, S, Rahnamoun, A, Reverdy, C, Errasti, G, Delacroix, T. | | 登録日 | 2023-11-19 | | 公開日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Computationally Driven Discovery and Characterization of SIRT3 Activating Compounds that Fully Recover Catalytic Activity under NAD+ Depletion
biorxiv, 2023
|
|
8V2N
 
 | | Human SIRT3 co-crystallized with ligands, including p53-AMC peptide and Carba-NAD | | 分子名称: | CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, GLN-PRO-LYS-FDL, NAD-dependent protein deacetylase sirtuin-3, ... | | 著者 | Chakrabarti, R, Ghosh, A, Guan, X, Upadhyay, A, Dumpati, R.K, Munshi, S, Roy, S, Chall, S, Rahnamoun, A, Reverdy, C, Errasti, G, Delacroix, T. | | 登録日 | 2023-11-23 | | 公開日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Computationally Driven Discovery and Characterization of SIRT3 Activating Compounds that Fully Recover Catalytic Activity under NAD+ Depletion
biorxiv, 2023
|
|
5CPR
 
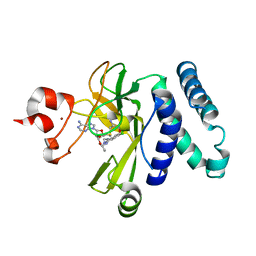 | | The novel SUV4-20 inhibitor A-196 verifies a role for epigenetics in genomic integrity | | 分子名称: | 6,7-dichloro-N-cyclopentyl-4-(pyridin-4-yl)phthalazin-1-amine, Histone-lysine N-methyltransferase SUV420H1, S-ADENOSYLMETHIONINE, ... | | 著者 | Jakob, C.G, Upadhyay, A.K, Sun, C. | | 登録日 | 2015-07-21 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | The SUV4-20 inhibitor A-196 verifies a role for epigenetics in genomic integrity.
Nat. Chem. Biol., 13, 2017
|
|
5VAR
 
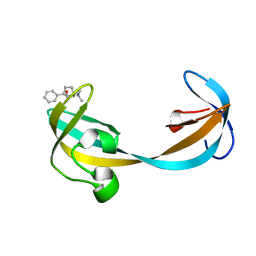 | |
2L4J
 
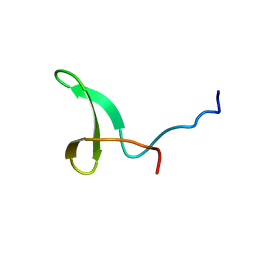 | | Yap ww2 | | 分子名称: | Yes-associated protein 2 (YAP2) | | 著者 | Webb, C, Upadhyay, A, Furutani-Seiki, M, Bagby, S. | | 登録日 | 2010-10-07 | | 公開日 | 2010-11-03 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Features and Ligand Binding Properties of Tandem WW Domains from YAP and TAZ, Nuclear Effectors of the Hippo Pathway.
Biochemistry, 50, 2011
|
|
2JOL
 
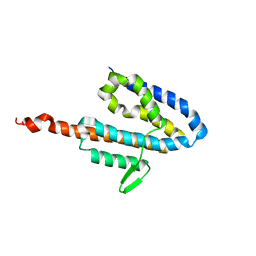 | | Average NMR structure of the catalytic domain of guanine nucleotide exchange factor BopE from Burkholderia pseudomallei | | 分子名称: | Putative G-nucleotide exchange factor | | 著者 | Wu, H, Upadhyay, A, Williams, C, Galyov, E.E, van den Elsen, J.M.H, Bagby, S. | | 登録日 | 2007-03-14 | | 公開日 | 2007-03-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The guanine-nucleotide-exchange factor BopE from Burkholderia pseudomallei adopts a compact version of the Salmonella SopE/SopE2 fold and undergoes a closed-to-open conformational change upon interaction with Cdc42
Biochem.J., 411, 2008
|
|
2JOK
 
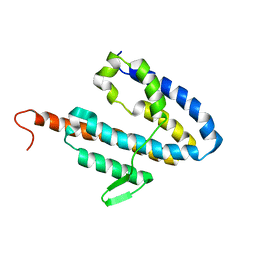 | | NMR structure of the catalytic domain of guanine nucleotide exchange factor BopE from Burkholderia pseudomallei | | 分子名称: | Putative G-nucleotide exchange factor | | 著者 | Wu, H, Upadhyay, A, Williams, C, Galyov, E.E, van den Elsen, J.M.H, Bagby, S. | | 登録日 | 2007-03-14 | | 公開日 | 2007-09-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The guanine-nucleotide-exchange factor BopE from Burkholderia pseudomallei adopts a compact version of the Salmonella SopE/SopE2 fold and undergoes a closed-to-open conformational change upon interaction with Cdc42
Biochem.J., 411, 2008
|
|
3KV5
 
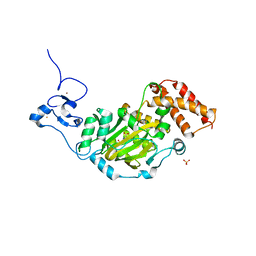 | | Structure of KIAA1718, human Jumonji demethylase, in complex with N-oxalylglycine | | 分子名称: | FE (II) ION, JmjC domain-containing histone demethylation protein 1D, N-OXALYLGLYCINE, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KV9
 
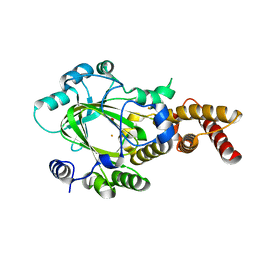 | | Structure of KIAA1718 Jumonji domain | | 分子名称: | FE (II) ION, JmjC domain-containing histone demethylation protein 1D, OXYGEN MOLECULE | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KVA
 
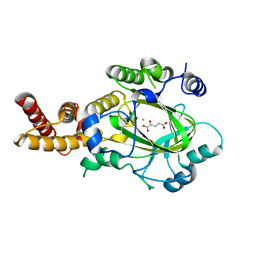 | | Structure of KIAA1718 Jumonji domain in complex with alpha-ketoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, FE (II) ION, JmjC domain-containing histone demethylation protein 1D, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KV6
 
 | | Structure of KIAA1718, human Jumonji demethylase, in complex with alpha-ketoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, FE (II) ION, JmjC domain-containing histone demethylation protein 1D, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KVB
 
 | | Structure of KIAA1718 Jumonji domain in complex with N-oxalylglycine | | 分子名称: | JmjC domain-containing histone demethylation protein 1D, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3PTR
 
 | | PHF2 Jumonji domain | | 分子名称: | 1,2-ETHANEDIOL, PHD finger protein 2 | | 著者 | Horton, J.R, Upadhyay, A.K, Hashimoto, H, Zhang, X, Cheng, X. | | 登録日 | 2010-12-03 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.954 Å) | | 主引用文献 | Structural basis for human PHF2 Jumonji domain interaction with metal ions.
J.Mol.Biol., 406, 2011
|
|
3KV4
 
 | | Structure of PHF8 in complex with histone H3 | | 分子名称: | 1,2-ETHANEDIOL, FE (II) ION, Histone H3-like, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Qi, H.H, Zhang, X, Shi, Y, Cheng, X. | | 登録日 | 2009-11-29 | | 公開日 | 2009-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3OS5
 
 | | SET7/9-Dnmt1 K142me1 complex | | 分子名称: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Dnmt1, ... | | 著者 | Esteve, P.-O, Chang, Y, Samaranayake, M, Upadhyay, A.K, Horton, J.R, Feehery, G.R, Cheng, X, Pradhan, S. | | 登録日 | 2010-09-08 | | 公開日 | 2010-12-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3RDH
 
 | | X-ray induced covalent inhibition of 14-3-3 | | 分子名称: | 14-3-3 protein zeta/delta, 4-[(E)-{4-formyl-5-hydroxy-6-methyl-3-[(phosphonooxy)methyl]pyridin-2-yl}diazenyl]benzoic acid, NICKEL (II) ION | | 著者 | Horton, J.R, Upadhyay, A.K, Fu, H, Cheng, X. | | 登録日 | 2011-04-01 | | 公開日 | 2011-09-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Discovery and structural characterization of a small molecule 14-3-3 protein-protein interaction inhibitor.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PUA
 
 | | PHF2 Jumonji-NOG-Ni(II) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, N-OXALYLGLYCINE, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Hashimoto, H, Zhang, X, Cheng, X. | | 登録日 | 2010-12-03 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Structural basis for human PHF2 Jumonji domain interaction with metal ions.
J.Mol.Biol., 406, 2011
|
|
3PUS
 
 | | PHF2 Jumonji-NOG-Ni(II) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, N-OXALYLGLYCINE, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Hashimoto, H, Zhang, X, Cheng, X. | | 登録日 | 2010-12-06 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Structural basis for human PHF2 Jumonji domain interaction with metal ions.
J.Mol.Biol., 406, 2011
|
|
