7ZZ4
 
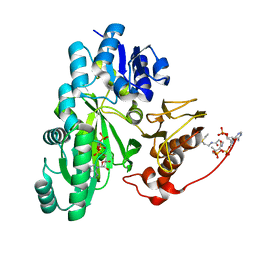 | | Cryo-EM structure of "BC closed" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZZ6
 
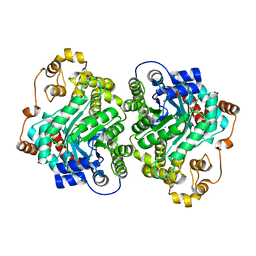 | | Cryo-EM structure of "CT-CT dimer" of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, PYRUVIC ACID, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
1JQ7
 
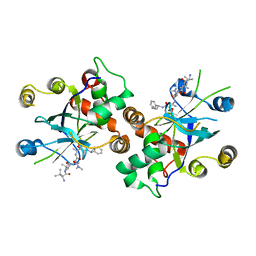 | | HCMV protease dimer-interface mutant, S225Y complexed to Inhibitor BILC 408 | | Descriptor: | ASSEMBLIN, N-(6-aminohexanoyl)-3-methyl-L-valyl-3-methyl-L-valyl-N~1~-[(2S,3S)-3-hydroxy-4-oxo-4-{[(1R)-1-phenylpropyl]amino}butan-2-yl]-N~4~,N~4~-dimethyl-L-aspartamide | | Authors: | Batra, R, Khayat, R, Tong, L. | | Deposit date: | 2001-08-03 | | Release date: | 2001-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular mechanism for dimerization to regulate the catalytic activity of human cytomegalovirus protease.
Nat.Struct.Biol., 8, 2001
|
|
5IKL
 
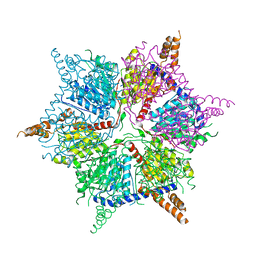 | |
4XYK
 
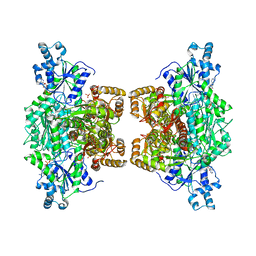 | | Crystal structure of human phosphofructokinase-1 in complex with ADP, Northeast Structural Genomics Consortium Target HR9275 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent 6-phosphofructokinase, platelet type, ... | | Authors: | Forouhar, F, Webb, B.A, Szu, F.-E, Seetharaman, J, Barber, D.L, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-02-02 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of human phosphofructokinase-1 and atomic basis of cancer-associated mutations.
Nature, 523, 2015
|
|
8G1U
 
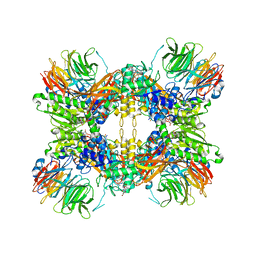 | | Structure of the methylosome-Lsm10/11 complex | | Descriptor: | ADENOSINE, Methylosome protein 50, Methylosome subunit pICln, ... | | Authors: | Lin, M, Paige, A, Tong, L. | | Deposit date: | 2023-02-03 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | In vitro methylation of the U7 snRNP subunits Lsm11 and SmE by the PRMT5/MEP50/pICln methylosome.
Rna, 29, 2023
|
|
8GXM
 
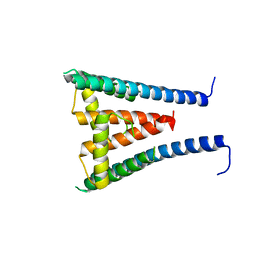 | | HUMAN SUGP1 433-586 | | Descriptor: | SURP and G-patch domain-containing protein 1 | | Authors: | Xu, K, Tong, L. | | Deposit date: | 2022-09-20 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | DHX15 is involved in SUGP1-mediated RNA missplicing by mutant SF3B1 in cancer.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8GXL
 
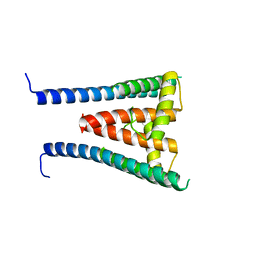 | | HUMAN SUGP1 433-577 | | Descriptor: | SURP and G-patch domain-containing protein 1 | | Authors: | Xu, K, Tong, L. | | Deposit date: | 2022-09-20 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | DHX15 is involved in SUGP1-mediated RNA missplicing by mutant SF3B1 in cancer.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5KS7
 
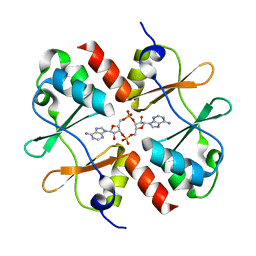 | | Crystal structure of Listeria monocytogenes OpuCA CBS domain dimer in complex with cyclic-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Carnitine transport ATP-binding protein OpuCA | | Authors: | Choi, P.H, Tong, L. | | Deposit date: | 2016-07-07 | | Release date: | 2016-07-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cyclic di-AMP targets the cystathionine beta-synthase domain of the osmolyte transporter OpuC.
Mol.Microbiol., 102, 2016
|
|
5KKN
 
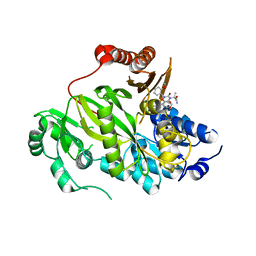 | | Crystal structure of human ACC2 BC domain in complex with ND-646, the primary amide of ND-630 | | Descriptor: | 2-[1-[(2~{R})-2-(2-methoxyphenyl)-2-(oxan-4-yloxy)ethyl]-5-methyl-6-(1,3-oxazol-2-yl)-2,4-bis(oxidanylidene)thieno[2,3-d]pyrimidin-3-yl]-2-methyl-propanamide, Acetyl-CoA carboxylase 2 | | Authors: | Wang, R, Paul, D, Tong, L. | | Deposit date: | 2016-06-22 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Acetyl-CoA carboxylase inhibition by ND-630 reduces hepatic steatosis, improves insulin sensitivity, and modulates dyslipidemia in rats.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5KXF
 
 | |
1H2H
 
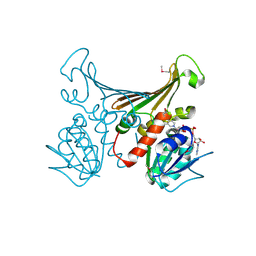 | | Crystal structure of TM1643 | | Descriptor: | HYPOTHETICAL PROTEIN TM1643, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yang, Z, Savchenko, A, Edwards, A, Arrowsmith, C, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-08-08 | | Release date: | 2002-08-15 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Aspartate dehydrogenase, a novel enzyme identified from structural and functional studies of TM1643.
J. Biol. Chem., 278, 2003
|
|
6UN9
 
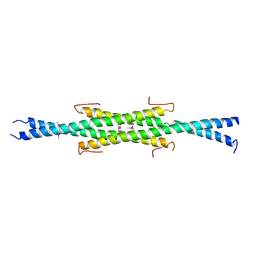 | | Crystal Structure of the Q7VLF5_HAEDU protein from Haemophilus ducreyi. Northeast Structural Genomics Consortium Target Hdr25 | | Descriptor: | Uncharacterized protein | | Authors: | Vorobiev, S.M, Seetharaman, J, Kolev, M, Xiao, R, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2019-10-11 | | Release date: | 2020-12-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Q7VLF5_HAEDU protein from Haemophilus ducreyi. Northeast Structural Genomics Consortium Target Hdr25
To Be Published
|
|
6URG
 
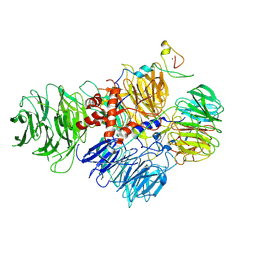 | | Cryo-EM structure of human CPSF160-WDR33-CPSF30-CPSF100 PIM complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 2, Cleavage and polyadenylation specificity factor subunit 4, ... | | Authors: | Sun, Y, Zhang, Y, Walz, T, Tong, L. | | Deposit date: | 2019-10-23 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural Insights into the Human Pre-mRNA 3'-End Processing Machinery.
Mol.Cell, 77, 2020
|
|
6URO
 
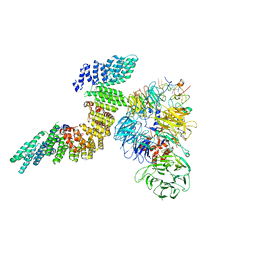 | | Cryo-EM structure of human CPSF160-WDR33-CPSF30-PAS RNA-CstF77 complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, Cleavage stimulation factor subunit 3, ... | | Authors: | Sun, Y, Zhang, Y, Walz, T, Tong, L. | | Deposit date: | 2019-10-23 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Insights into the Human Pre-mRNA 3'-End Processing Machinery.
Mol.Cell, 77, 2020
|
|
5VYZ
 
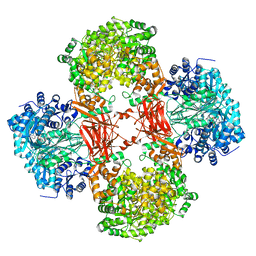 | | Crystal structure of Lactococcus lactis pyruvate carboxylase in complex with cyclic-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Choi, P.H, Tong, L. | | Deposit date: | 2017-05-26 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional studies of pyruvate carboxylase regulation by cyclic di-AMP in lactic acid bacteria.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VZ0
 
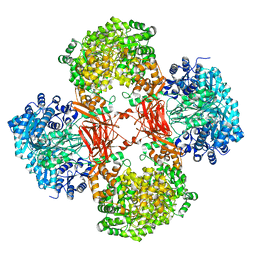 | | Crystal structure of Lactococcus lactis pyruvate carboxylase G746A mutant in complex with cyclic-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Choi, P.H, Tong, L. | | Deposit date: | 2017-05-26 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional studies of pyruvate carboxylase regulation by cyclic di-AMP in lactic acid bacteria.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6E40
 
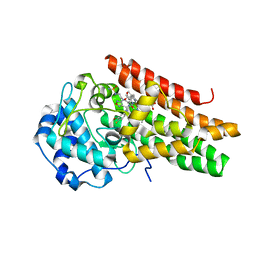 | | Crystal structure of the indoleamine 2,3-dioxygenase 1 (IDO1) in complexed with ferric heme and Epacadostat | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-(3-bromo-4-fluorophenyl)-N'-hydroxy-4-{[2-(sulfamoylamino)ethyl]amino}-1,2,5-oxadiazole-3-carboximidamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6E42
 
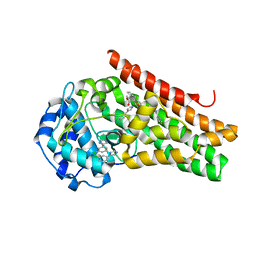 | | CRYSTAL STRUCTURE OF HUMAN INDOLEAMINE 2,3-DIOXYGENASE 1 (IDO1) in complex with ferric heme and 4-Chlorophenyl imidazole | | Descriptor: | 4-(3-chlorophenyl)-1H-imidazole, Indoleamine 2,3-dioxygenase 1, PHOSPHATE ION, ... | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6E41
 
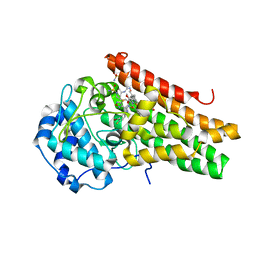 | | CRYSTAL STRUCTURE OF HUMAN INDOLEAMINE 2,3-DIOXYGENASE 1 (IDO1) in complex with ferric heme and an Epacadostat analog | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-(3-bromo-4-fluorophenyl)-N'-hydroxy-4-{[2-(sulfamoylamino)ethyl]sulfanyl}-1,2,5-oxadiazole-3-carboximidamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6E43
 
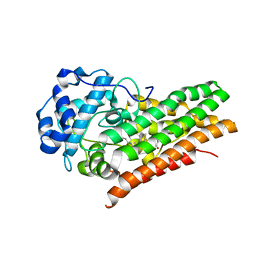 | | Crystal structure of human indoleamine 2,3-dioxygenase 1 (IDO1) in complex with a BMS-978587 analog | | Descriptor: | (1R,2S)-2-(4-[cyclohexyl(2-methylpropyl)amino]-3-{[(4-methylphenyl)carbamoyl]amino}phenyl)cyclopropane-1-carboxylic acid, BENZOIC ACID, Indoleamine 2,3-dioxygenase 1 | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6E45
 
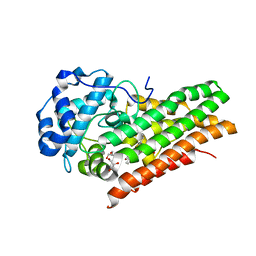 | | CRYSTAL STRUCTURE OF HUMAN INDOLEAMINE 2,3-DIOXYGENASE 1 (IDO1) free enzyme in the ferrous state | | Descriptor: | GLYCEROL, Indoleamine 2,3-dioxygenase 1, PHOSPHATE ION, ... | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6E44
 
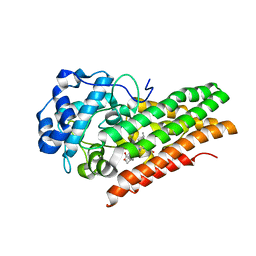 | | CRYSTAL STRUCTURE OF HUMAN INDOLEAMINE 2,3-DIOXYGENASE 1 (IDO1) free enzyme in the ferric state | | Descriptor: | Indoleamine 2,3-dioxygenase 1, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6E46
 
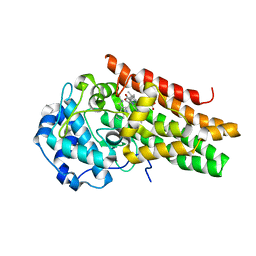 | | CRYSTAL STRUCTURE OF HUMAN INDOLEAMINE 2,3-DIOXYGENASE 1 (IDO1) in complex with ferrous heme and tryptophan | | Descriptor: | Indoleamine 2,3-dioxygenase 1, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2018-07-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | High-resolution structures of inhibitor complexes of human indoleamine 2,3-dioxygenase 1 in a new crystal form.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
1G3N
 
 | |
