5MDX
 
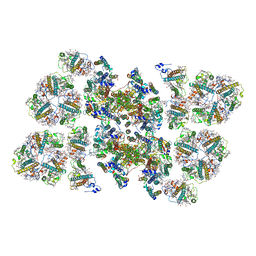 | | Cryo-EM structure of the PSII supercomplex from Arabidopsis thaliana | | Descriptor: | CHLOROPHYLL A, CHLOROPHYLL B, Chlorophyll a-b binding protein 1, ... | | Authors: | van Bezouwen, L.S, Caffarri, S, Kale, R.S, Kouril, R, Thunnissen, A.M.W.H, Oostergetel, G.T, Boekema, E.J. | | Deposit date: | 2016-11-13 | | Release date: | 2017-06-21 | | Last modified: | 2019-04-24 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Subunit and chlorophyll organization of the plant photosystem II supercomplex.
Nat Plants, 3, 2017
|
|
5OFQ
 
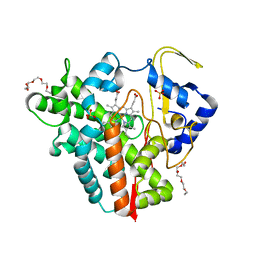 | |
7B4J
 
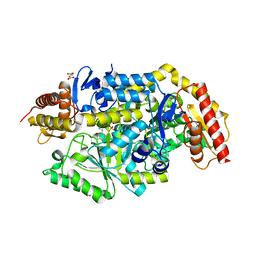 | | Thermostable omega transaminase PjTA-R6 variant W58M/F86L/R417L engineered for asymmetric synthesis of enantiopure bulky amines | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aspartate aminotransferase family protein, SUCCINIC ACID | | Authors: | Capra, N, Rozeboom, H.J, Thunnissen, A.M.W.H, Janssen, D.B. | | Deposit date: | 2020-12-02 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational Redesign of an omega-Transaminase from Pseudomonas jessenii for Asymmetric Synthesis of Enantiopure Bulky Amines.
Acs Catalysis, 11, 2021
|
|
7B4I
 
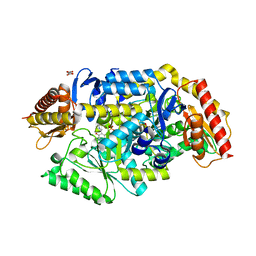 | | Thermostable omega transaminase PjTA-R6 variant W58G engineered for asymmetric synthesis of enantiopure bulky amines | | Descriptor: | Aspartate aminotransferase family protein, PYRIDOXAL-5'-PHOSPHATE, SUCCINIC ACID | | Authors: | Capra, N, Rozeboom, H.J, Thunnissen, A.M.W.H, Janssen, D.B. | | Deposit date: | 2020-12-02 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational Redesign of an omega-Transaminase from Pseudomonas jessenii for Asymmetric Synthesis of Enantiopure Bulky Amines.
Acs Catalysis, 11, 2021
|
|
6I8N
 
 | |
1CKP
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR PURVALANOL B | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN (CYCLIN-DEPENDENT PROTEIN KINASE 2), PURVALANOL B | | Authors: | Gray, N.S, Thunnissen, A.M.W.H, Schultz, P.G, Kim, S.H. | | Deposit date: | 1998-07-14 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors.
Science, 281, 1998
|
|
6GK6
 
 | |
4U5P
 
 | | Crystal structure of native RhCC (YP_702633.1) from Rhodococcus jostii RHA1 at 1.78 Angstrom | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Poddar, H, Rozeboom, H.J, Thunnissen, A.M.W.H. | | Deposit date: | 2014-07-25 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | Functional and structural characterization of an unusual cofactor-independent oxygenase.
Biochemistry, 54, 2015
|
|
4U5R
 
 | |
4ESF
 
 | |
4ESB
 
 | | Crystal structure of PadR-like transcriptional regulator (BC4206) from Bacillus cereus strain ATCC 14579 | | Descriptor: | GLYCEROL, SULFATE ION, Transcriptional regulator, ... | | Authors: | Fibriansah, G, Kovacs, A.T, Kuipers, O.P, Thunnissen, A.M.W.H. | | Deposit date: | 2012-04-23 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Two Transcriptional Regulators from Bacillus cereus Define the Conserved Structural Features of a PadR Subfamily.
Plos One, 7, 2012
|
|
2AE0
 
 | | Crystal structure of MltA from Escherichia coli reveals a unique lytic transglycosylase fold | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Membrane-bound lytic murein transglycosylase A | | Authors: | Van Straaten, K.E, Dijkstra, B.W, Vollmer, W, Thunnissen, A.M.W.H. | | Deposit date: | 2005-07-21 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of MltA from Escherichia coli Reveals a Unique Lytic Transglycosylase Fold
J.Mol.Biol., 352, 2005
|
|
3L6H
 
 | | Crystal structure of lactococcal OpuAC in its closed-liganded conformation complexed with glycine betaine | | Descriptor: | Betaine ABC transporter permease and substrate binding protein, CHLORIDE ION, TRIMETHYL GLYCINE | | Authors: | Berntsson, R.P.A, Wolters, J.C, Gul, N, Karasawa, A, Thunnissen, A.M.W.H, Slotboom, D.J, Poolman, B. | | Deposit date: | 2009-12-23 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand binding and crystal structures of the substrate-binding domain of the ABC transporter OpuA.
Plos One, 5, 2010
|
|
3L6G
 
 | | Crystal structure of lactococcal OpuAC in its open conformation | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Betaine ABC transporter permease and substrate binding protein | | Authors: | Berntsson, R.P.A, Wolters, J.C, Gul, N, Karasawa, A, Thunnissen, A.M.W.H, Slotboom, D.J, Poolman, B. | | Deposit date: | 2009-12-23 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand binding and crystal structures of the substrate-binding domain of the ABC transporter OpuA.
Plos One, 5, 2010
|
|
8BIT
 
 | | Crystal structure of acyl-CoA synthetase from Metallosphaera sedula in complex with Coenzyme A and acetyl-AMP | | Descriptor: | 4-hydroxybutyrate--CoA ligase 1, COENZYME A, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] ethanoate | | Authors: | Capra, N, Thunnissen, A.M.W.H, Janssen, D.B. | | Deposit date: | 2022-11-02 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Adapting an acyl CoA ligase from Metallosphaera sedula for lactam formation by structure-guided protein engineering
Front Catal, 4, 2024
|
|
8BIQ
 
 | | Crystal structure of acyl-COA synthetase from Metallosphaera sedula in complex with acetyl-AMP | | Descriptor: | 4-hydroxybutyrate--CoA ligase 1, ADENOSINE MONOPHOSPHATE, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] ethanoate | | Authors: | Capra, N, Thunnissen, A.M.W.H, Janssen, D.B. | | Deposit date: | 2022-11-02 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Adapting an acyl CoA ligase from Metallosphaera sedula for lactam formation by structure-guided protein engineering
Front Catal, 4, 2024
|
|
6S9V
 
 | | Crystal structure of sucrose 6F-phosphate phosphorylase from Thermoanaerobacter thermosaccharolyticum | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Capra, N, Franceus, J, Desmet, T, Thunnissen, A.M.W.H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Comparison of a Promiscuous and a Highly Specific Sucrose 6 F -Phosphate Phosphorylase.
Int J Mol Sci, 20, 2019
|
|
6RX8
 
 | |
6RXA
 
 | | EDDS lyase variant D290M/Y320M with bound formate | | Descriptor: | Argininosuccinate lyase, FORMIC ACID, GLYCEROL, ... | | Authors: | Grandi, E, Poelarends, G.J, Thunnissen, A.M.W.H. | | Deposit date: | 2019-06-07 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Engineered C-N Lyase: Enantioselective Synthesis of Chiral Synthons for Artificial Dipeptide Sweeteners.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6S9U
 
 | | Crystal structure of sucrose 6F-phosphate phosphorylase from Ilumatobacter coccineus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, ... | | Authors: | Capra, N, Franceus, J, Desmet, T, Thunnissen, A.M.W.H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Comparison of a Promiscuous and a Highly Specific Sucrose 6 F -Phosphate Phosphorylase.
Int J Mol Sci, 20, 2019
|
|
2PIC
 
 | |
2PI8
 
 | | Crystal structure of E. coli MltA with bound chitohexaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Membrane-bound lytic murein transglycosylase A, PHOSPHATE ION | | Authors: | van Straaten, K.E, Barends, T.R.M, Dijkstra, B.W, Thunnissen, A.M.W.H. | | Deposit date: | 2007-04-13 | | Release date: | 2007-05-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Escherichia coli Lytic transglycosylase MltA with bound chitohexaose: implications for peptidoglycan binding and cleavage
J.Biol.Chem., 282, 2007
|
|
2PJJ
 
 | |
5L94
 
 | |
5L91
 
 | |
