8EOP
 
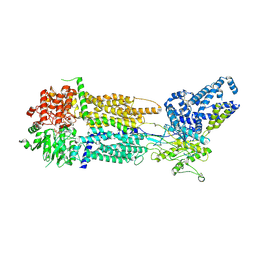 | | Cryo-EM Structure of Nanodisc reconstituted human ABCA7 EQ mutant in ATP bound closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Alam, A, Le, L.T.M, Thompson, J.R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of human ABCA7 provide insights into its phospholipid translocation mechanisms.
Embo J., 42, 2023
|
|
8EE6
 
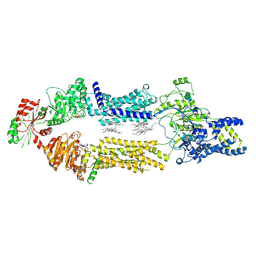 | | Cryo-EM Structure of human ABCA7 in PE/Ch nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Phospholipid-transporting ATPase ABCA7, ... | | Authors: | Alam, A, Le, L.T.M, Thompson, J.R. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of human ABCA7 provide insights into its phospholipid translocation mechanisms.
Embo J., 42, 2023
|
|
8EDW
 
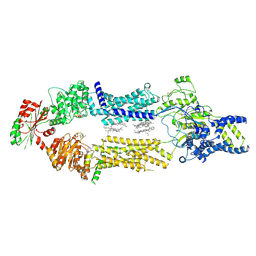 | | Cryo-EM Structure of human ABCA7 in BPL/Ch Nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Alam, A, Le, L.T.M, Thompson, J.R. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of human ABCA7 provide insights into its phospholipid translocation mechanisms.
Embo J., 42, 2023
|
|
3L1X
 
 | |
3L1Y
 
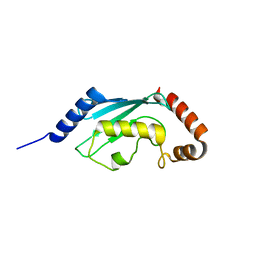 | |
3SQF
 
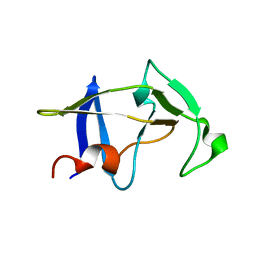 | | Crystal structure of monomeric M-PMV retroviral protease | | Descriptor: | Protease | | Authors: | Jaskolski, M, Kazmierczyk, M, Gilski, M, Krzywda, S, Pichova, I, Zabranska, H, Khatib, F, DiMaio, F, Cooper, S, Thompson, J, Popovic, Z, Baker, D, Group, Foldit Contenders | | Deposit date: | 2011-07-05 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6324 Å) | | Cite: | Crystal structure of a monomeric retroviral protease solved by protein folding game players.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6CO2
 
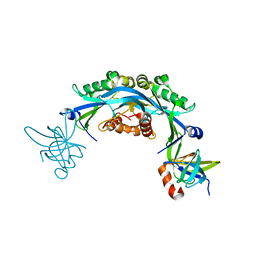 | | Structure of an engineered protein (NUDT16TI) in complex with 53BP1 Tudor domains | | Descriptor: | NUDT16-Tudor-interacting (NUDT16TI), TP53-binding protein 1 | | Authors: | Botuyan, M.V, Thompson, J.R, Cui, G, Mer, G. | | Deposit date: | 2018-03-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3L1Z
 
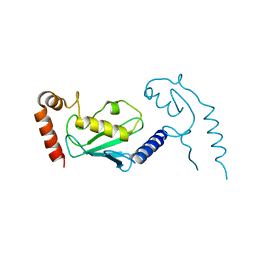 | |
1EZF
 
 | | CRYSTAL STRUCTURE OF HUMAN SQUALENE SYNTHASE | | Descriptor: | FARNESYL-DIPHOSPHATE FARNESYLTRANSFERASE, N-{2-[TRANS-7-CHLORO-1-(2,2-DIMETHYL-PROPYL) -5-NAPHTHALEN-1-YL-2-OXO-1,2,3,5-TETRAHYDRO-BENZO[E] [1,4]OXAZEPIN-3-YL]-ACETYL}-ASPARTIC ACID | | Authors: | Pandit, J, Danley, D.E, Schulte, G.K, Mazzalupo, S.M, Pauly, T.A, Hayward, C.M, Hamanaka, E.S, Thompson, J.F, Harwood, H.J. | | Deposit date: | 2000-05-10 | | Release date: | 2000-10-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of human squalene synthase. A key enzyme in cholesterol biosynthesis.
J.Biol.Chem., 275, 2000
|
|
3KTF
 
 | |
4W8Q
 
 | |
3SZM
 
 | |
3T1N
 
 | |
8U4U
 
 | |
3U79
 
 | | AL-103 Y32F Y96F | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, Amyloidogenic immunoglobulin light chain protein AL-103 Y32F Y96F, ... | | Authors: | DiCostanzo, A.C, Thompson, J.R, Ramirez-Alvarado, M. | | Deposit date: | 2011-10-13 | | Release date: | 2012-07-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Tyrosine Residues mediate crucial interactions in amyloid formation for immunoglobulin light chains
To be Published
|
|
3U3Z
 
 | | Structure of human microcephalin (MCPH1) tandem BRCT domains in complex with an H2A.X peptide phosphorylated at Ser139 and Tyr142 | | Descriptor: | GLYCEROL, Histone H2A.X peptide, Microcephalin | | Authors: | Singh, N, Thompson, J.R, Heroux, A, Mer, G. | | Deposit date: | 2011-10-06 | | Release date: | 2012-07-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Dual recognition of phosphoserine and phosphotyrosine in histone variant H2A.X by DNA damage response protein MCPH1.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1DC9
 
 | | PROPERTIES AND CRYSTAL STRUCTURE OF A BETA-BARREL FOLDING MUTANT, V60N INTESTINAL FATTY ACID BINDING PROTEIN (IFABP) | | Descriptor: | INTESTINAL FATTY ACID BINDING PROTEIN | | Authors: | Ropson, I.J, Yowler, B.C, Dalessio, P.M, Banaszak, L, Thompson, J. | | Deposit date: | 1999-11-04 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Properties and crystal structure of a beta-barrel folding mutant.
Biophys.J., 78, 2000
|
|
1IB6
 
 | | CRYSTAL STRUCTURE OF R153C E. COLI MALATE DEHYDROGENASE | | Descriptor: | MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Bell, J.K, Yennawar, H.P, Wright, S.K, Thompson, J.R, Viola, R.E, Banaszak, L.J. | | Deposit date: | 2001-03-27 | | Release date: | 2001-09-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analyses of a Malate Dehydrogenase with a Variable Active Site
J.Biol.Chem., 276, 2001
|
|
1IE3
 
 | | CRYSTAL STRUCTURE OF R153C E. COLI MALATE DEHYDROGENASE | | Descriptor: | MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PYRUVIC ACID | | Authors: | Bell, J.K, Yennawar, H.P, Wright, S.K, Thompson, J.R, Viola, R.E, Banaszak, L.J. | | Deposit date: | 2001-04-05 | | Release date: | 2001-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analyses of a Malate Dehydrogenase with a Variable Active Site
J.Biol.Chem., 276, 2001
|
|
3TVV
 
 | |
3U7A
 
 | | AL-09 Y32F Y96F | | Descriptor: | Amyloidogenic immunoglobulin light chain protein AL-09 Y32F Y96F, variable domain | | Authors: | DiCostanzo, A.C, Thompson, J.R, Ramirez-Alvarado, M. | | Deposit date: | 2011-10-13 | | Release date: | 2012-07-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tyrosine Residues mediate crucial interactions in amyloid formation for immunoglobulin light chains
To be Published
|
|
3TW1
 
 | | Structure of Rtt106-AHN | | Descriptor: | GLYCEROL, Histone chaperone RTT106, N-[2-(1H-IMIDAZOL-4-YL)ETHYL]ACETAMIDE | | Authors: | Su, D, Thompson, J.R, Mer, G. | | Deposit date: | 2011-09-21 | | Release date: | 2012-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | Structural basis for recognition of H3K56-acetylated histone H3-H4 by the chaperone Rtt106.
Nature, 483, 2012
|
|
5TIH
 
 | | Structural basis for inhibition of erythrocyte invasion by antibodies to Plasmodium falciparum protein CyRPA | | Descriptor: | ACETATE ION, CyRPA antibody Fab Heavy Chain, CyRPA antibody Fab Light Chain, ... | | Authors: | Chen, L, Xu, Y, Wang, W, Thompson, J.K, Goddard-Borger, E, Lawrence, M.C, Cowman, A.F. | | Deposit date: | 2016-10-03 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural basis for inhibition of erythrocyte invasion by antibodies toPlasmodium falciparumprotein CyRPA.
Elife, 6, 2017
|
|
5TIK
 
 | | Structural basis for inhibition of erythrocyte invasion by antibodies to Plasmodium falciparum protein CyRPA | | Descriptor: | Cysteine-rich protective antigen | | Authors: | Chen, L, Xu, Y, Wang, W, Thompson, J.K, Goddard-Borger, E, Lawrence, M.C, Cowman, A.F. | | Deposit date: | 2016-10-03 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural basis for inhibition of erythrocyte invasion by antibodies toPlasmodium falciparumprotein CyRPA.
Elife, 6, 2017
|
|
5TRE
 
 | | Zinc and the Iron Donor Frataxin Regulate Oligomerization of the Scaffold Protein to Form New Fe-S Cluster Assembly Centers | | Descriptor: | Frataxin homolog, mitochondrial, Iron sulfur cluster assembly protein 1 | | Authors: | Ranatunga, W, Gakh, O, Galeano, B.K, Smith IV, D.Y, Thompson, J.R, Isaya, G. | | Deposit date: | 2016-10-26 | | Release date: | 2017-06-07 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (15.6 Å) | | Cite: | Zinc and the iron donor frataxin regulate oligomerization of the scaffold protein to form new Fe-S cluster assembly centers.
Metallomics, 9, 2017
|
|
