4V7H
 
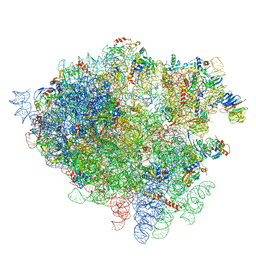 | | Structure of the 80S rRNA and proteins and P/E tRNA for eukaryotic ribosome based on cryo-EM map of Thermomyces lanuginosus ribosome at 8.9A resolution | | 分子名称: | 18S rRNA, 26S ribosomal RNA, 40S ribosomal protein S0(A), ... | | 著者 | Taylor, D.J, Devkota, B, Huang, A.D, Topf, M, Narayanan, E, Sali, A, Harvey, S.C, Frank, J. | | 登録日 | 2009-09-22 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (8.9 Å) | | 主引用文献 | Comprehensive molecular structure of the eukaryotic ribosome.
Structure, 17, 2009
|
|
7R6Z
 
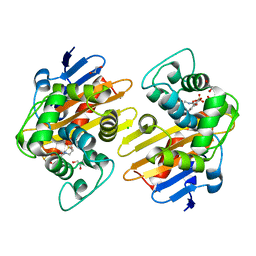 | | OXA-48 bound by Compound 3.3 | | 分子名称: | 1,2-ETHANEDIOL, 4-amino-5-hydroxynaphthalene-2,7-disulfonic acid, Beta-lactamase, ... | | 著者 | Taylor, D.M, Hu, L, Prasad, B.V.V, Sankaran, B, Palzkill, T. | | 登録日 | 2021-06-24 | | 公開日 | 2021-12-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
6XQR
 
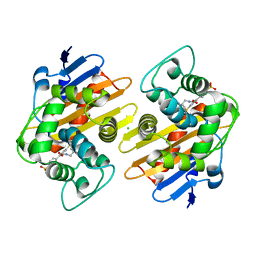 | | OXA-48 bound by Compound 2.2 | | 分子名称: | Beta-lactamase, CHLORIDE ION, [1,1'-biphenyl]-4,4'-disulfonic acid | | 著者 | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | 登録日 | 2020-07-10 | | 公開日 | 2021-12-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
6UVK
 
 | | OXA-48 bound by inhibitor CDD-97 | | 分子名称: | 1,2-ETHANEDIOL, 1-{4-[4-(2-ethoxyphenyl)piperazin-1-yl]-1,3,5-triazin-2-yl}piperidine-4-carboxylic acid, Beta-lactamase, ... | | 著者 | Taylor, D.M, Hu, L, Prasad, B.V.V, Sankaran, B, Palzkill, T.G. | | 登録日 | 2019-11-02 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Identifying Oxacillinase-48 Carbapenemase Inhibitors Using DNA-Encoded Chemical Libraries.
Acs Infect Dis., 6, 2020
|
|
7L8O
 
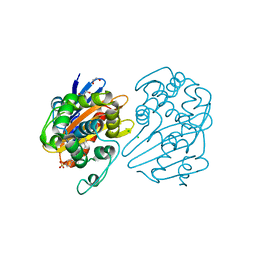 | | OXA-48 bound by Compound 4.3 | | 分子名称: | 1,2-ETHANEDIOL, 9H-fluorene-2,7-disulfonate, Beta-lactamase, ... | | 著者 | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | 登録日 | 2020-12-31 | | 公開日 | 2021-12-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
7K5V
 
 | | OXA-48 bound by Compound 3.1 | | 分子名称: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | 著者 | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | 登録日 | 2020-09-17 | | 公開日 | 2021-12-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
2P8Y
 
 | | Fitted structure of ADPR-eEF2 in the 80S:ADPR-eEF2:GDP:sordarin cryo-EM reconstruction | | 分子名称: | ADENOSINE-5-DIPHOSPHORIBOSE, Elongation factor 2, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Taylor, D.J, Nilsson, J, Merrill, A.R, Andersen, G.R, Nissen, P, Frank, J. | | 登録日 | 2007-03-23 | | 公開日 | 2007-05-08 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (11.7 Å) | | 主引用文献 | Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation.
Embo J., 26, 2007
|
|
2P8Z
 
 | | Fitted structure of ADPR-eEF2 in the 80S:ADPR-eEF2:GDPNP:sordarin cryo-EM reconstruction | | 分子名称: | ADENOSINE-5-DIPHOSPHORIBOSE, Elongation factor 2, Elongation factor Tu-B, ... | | 著者 | Taylor, D.J, Nilsson, J, Merrill, A.R, Andersen, G.R, Nissen, P, Frank, J. | | 登録日 | 2007-03-23 | | 公開日 | 2007-05-08 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (8.9 Å) | | 主引用文献 | Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation.
Embo J., 26, 2007
|
|
2P8X
 
 | | Fitted structure of ADPR-eEF2 in the 80S:ADPR-eEF2:GDPNP cryo-EM reconstruction | | 分子名称: | ADENOSINE-5-DIPHOSPHORIBOSE, Elongation factor 2, Elongation factor Tu-B, ... | | 著者 | Taylor, D.J, Nilsson, J, Merrill, A.R, Andersen, G.R, Nissen, P, Frank, J. | | 登録日 | 2007-03-23 | | 公開日 | 2007-05-08 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (9.7 Å) | | 主引用文献 | Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation.
Embo J., 26, 2007
|
|
4CLN
 
 | | STRUCTURE OF A RECOMBINANT CALMODULIN FROM DROSOPHILA MELANOGASTER REFINED AT 2.2-ANGSTROMS RESOLUTION | | 分子名称: | CALCIUM ION, CALMODULIN | | 著者 | Taylor, D.A, Sack, J.S, Maune, J.F, Beckingham, K, Quiocho, F.A. | | 登録日 | 1991-06-24 | | 公開日 | 1992-07-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of a recombinant calmodulin from Drosophila melanogaster refined at 2.2-A resolution.
J.Biol.Chem., 266, 1991
|
|
2P8W
 
 | | Fitted structure of eEF2 in the 80S:eEF2:GDPNP cryo-EM reconstruction | | 分子名称: | Elongation factor 2, Elongation factor Tu-B, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | 著者 | Taylor, D.J, Nilsson, J, Merrill, A.R, Andersen, G.R, Nissen, P, Frank, J. | | 登録日 | 2007-03-23 | | 公開日 | 2007-05-08 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (11.3 Å) | | 主引用文献 | Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation.
Embo J., 26, 2007
|
|
7JHQ
 
 | | OXA-48 bound by Compound 2.3 | | 分子名称: | 1,2-ETHANEDIOL, Beta-lactamase OXA-48, CHLORIDE ION, ... | | 著者 | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | 登録日 | 2020-07-21 | | 公開日 | 2021-12-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
7N9T
 
 | |
6E53
 
 | | Structure of TERT in complex with a novel telomerase inhibitor | | 分子名称: | MAGNESIUM ION, RNA/DNA hairpin, Telomerase reverse transcriptase, ... | | 著者 | Hernandez-Sanchez, W, Huang, W, Plucinsky, B, Garcia-Vazquez, N, Berdis, A.J, Skordalakes, E, Taylor, D.J. | | 登録日 | 2018-07-19 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A non-natural nucleotide uses a specific pocket to selectively inhibit telomerase activity.
Plos Biol., 17, 2019
|
|
7S4X
 
 | | Cas9:gRNA in complex with 18-20MM DNA, 1 minute time-point, kinked active conformation | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, NTS, ... | | 著者 | Bravo, J.P.K, Taylor, D.W, Liu, M.S, Johnson, K.A. | | 登録日 | 2021-09-09 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.76 Å) | | 主引用文献 | Structural basis for mismatch surveillance by CRISPR-Cas9.
Nature, 603, 2022
|
|
7S4U
 
 | | Cryo-EM structure of Cas9 in complex with 12-14MM DNA substrate, 5 minute time-point | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, Non-target strand, Target strand, ... | | 著者 | Bravo, J.P.K, Taylor, D.W, Liu, M.S, Johnson, K.A. | | 登録日 | 2021-09-09 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.56 Å) | | 主引用文献 | Structural basis for mismatch surveillance by CRISPR-Cas9.
Nature, 603, 2022
|
|
7SBB
 
 | |
7SBA
 
 | |
7S9W
 
 | | Structure of DrmAB:ADP:DNA complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*T)-3'), DrmA, ... | | 著者 | Bravo, J.P.K, Taylor, D.W, Brounds, S.J.J, Aparicio-Maldonado, C. | | 登録日 | 2021-09-21 | | 公開日 | 2022-06-15 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis for broad anti-phage immunity by DISARM.
Nat Commun, 13, 2022
|
|
7S4V
 
 | | Cas9 bound to 12-14MM DNA, 60 min time-point, kinked conformation | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, NTS, TS, ... | | 著者 | Bravo, J.P.K, Taylor, D.W, Liu, M.S, Johnson, K.A. | | 登録日 | 2021-09-09 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | Structural basis for mismatch surveillance by CRISPR-Cas9.
Nature, 603, 2022
|
|
7S9V
 
 | | DrmAB:ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DrmA, DrmB | | 著者 | Bravo, J.P.K, Taylor, D.W, Brouns, S.J.J, Aparicio-Maldonado, C. | | 登録日 | 2021-09-21 | | 公開日 | 2022-06-15 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis for broad anti-phage immunity by DISARM.
Nat Commun, 13, 2022
|
|
8FZR
 
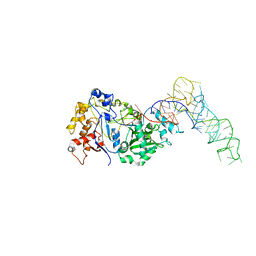 | |
8TL0
 
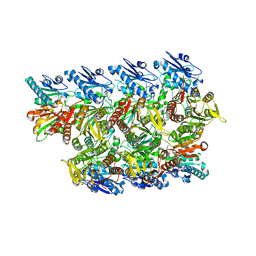 | |
7KOG
 
 | | Lethocerus Myosin II complete coiled-coil domain resolved in its native environment | | 分子名称: | Myosin heavy chain isoform Mhc_X1 | | 著者 | Rahmani, H, Hu, Z, Daneshparvar, N, Taylor, D, Taylor, K.A. | | 登録日 | 2020-11-09 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (4.25 Å) | | 主引用文献 | The myosin II coiled-coil domain atomic structure in its native environment.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8U3Y
 
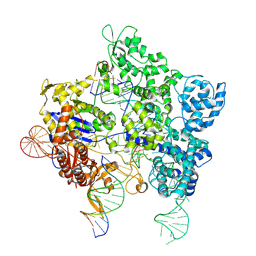 | | SpG Cas9 with NGG PAM DNA target | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(P*CP*GP*TP*TP*TP*GP*TP*AP*CP*TP*CP*CP*AP*GP*CP*G)-3'), DNA (5'-D(P*TP*CP*TP*CP*AP*TP*CP*TP*TP*TP*AP*TP*GP*CP*GP*TP*C)-3'), ... | | 著者 | Bravo, J.P.K, Hibshman, G.N, Taylor, D.W. | | 登録日 | 2023-09-08 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Unraveling the mechanisms of PAMless DNA interrogation by SpRY-Cas9.
Nat Commun, 15, 2024
|
|
