5NIM
 
 | | EthR complex | | Descriptor: | HTH-type transcriptional regulator EthR, SULFATE ION, [1-(2-hydroxyethyl)pyrrolo[3,4-c]pyrazol-5-yl]-(5-propyl-1,2-oxazol-3-yl)methanone | | Authors: | Pohl, E, Tatum, N.J, Cole, J.C, Baulard, A.R. | | Deposit date: | 2017-03-24 | | Release date: | 2017-11-15 | | Last modified: | 2017-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | New active leads for tuberculosis booster drugs by structure-based drug discovery.
Org. Biomol. Chem., 15, 2017
|
|
5NJ0
 
 | | EthR complex | | Descriptor: | HTH-type transcriptional regulator EthR, SULFATE ION, ~{N}-[(2~{R})-2-(4-nitrophenyl)-2-oxidanyl-ethyl]-1,3-benzodioxole-5-carboxamide | | Authors: | Pohl, E, Tatum, N.J, Cole, J.C, Baulard, A.R. | | Deposit date: | 2017-03-27 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New active leads for tuberculosis booster drugs by structure-based drug discovery.
Org. Biomol. Chem., 15, 2017
|
|
5NIO
 
 | | EthR complex | | Descriptor: | HTH-type transcriptional regulator EthR, SULFATE ION, ~{N}-butyl-4-methyl-piperidine-1-carboxamide | | Authors: | Pohl, E, Tatum, N.J, Cole, J.C, Baulard, A.R. | | Deposit date: | 2017-03-24 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | New active leads for tuberculosis booster drugs by structure-based drug discovery.
Org. Biomol. Chem., 15, 2017
|
|
5NIZ
 
 | | EthR complex | | Descriptor: | 1-[3-[5-[(1~{R},2~{R})-2-methylcyclopropyl]furan-2-yl]propanoyl]piperidine-4-carboxamide, HTH-type transcriptional regulator EthR, SULFATE ION | | Authors: | Pohl, E, Tatum, N.J, Cole, J.C, Baulard, A.R. | | Deposit date: | 2017-03-27 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New active leads for tuberculosis booster drugs by structure-based drug discovery.
Org. Biomol. Chem., 15, 2017
|
|
6GU6
 
 | | CDK1/Cks2 in complex with Dinaciclib | | Descriptor: | 3-[({3-ethyl-5-[(2S)-2-(2-hydroxyethyl)piperidin-1-yl]pyrazolo[1,5-a]pyrimidin-7-yl}amino)methyl]-1-hydroxypyridinium, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GU4
 
 | | CDK1/CyclinB/Cks2 in complex with CGP74514A | | Descriptor: | Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, G2/mitotic-specific cyclin-B1, ... | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GUH
 
 | | CDK2 in complex with AZD5438 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-methyl-3-propan-2-yl-imidazol-4-yl)-~{N}-(4-methylsulfonylphenyl)pyrimidin-2-amine, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GUC
 
 | | CDK2/CyclinA in complex with SU9516 | | Descriptor: | (3Z)-3-(1H-IMIDAZOL-5-YLMETHYLENE)-5-METHOXY-1H-INDOL-2(3H)-ONE, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GUK
 
 | | CDK2 in complex with CGP74514A | | Descriptor: | Cyclin-dependent kinase 2, ~{N}2-[(1~{R},2~{S})-2-azanylcyclohexyl]-~{N}6-(3-chlorophenyl)-9-ethyl-purine-2,6-diamine | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GUE
 
 | | CDK2/CyclinA in complex with AZD5438 | | Descriptor: | 4-(2-methyl-3-propan-2-yl-imidazol-4-yl)-~{N}-(4-methylsulfonylphenyl)pyrimidin-2-amine, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GU2
 
 | | CDK1/CyclinB/Cks2 in complex with Flavopiridol | | Descriptor: | 2-(2-chlorophenyl)-8-[(3~{R},4~{R})-1-methyl-3-oxidanyl-piperidin-4-yl]-5,7-bis(oxidanyl)chromen-4-one, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, ... | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GUB
 
 | | CDK2/CyclinA in complex with Flavopiridol | | Descriptor: | 2-(2-chlorophenyl)-8-[(3~{R},4~{R})-1-methyl-3-oxidanyl-piperidin-4-yl]-5,7-bis(oxidanyl)chromen-4-one, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GU3
 
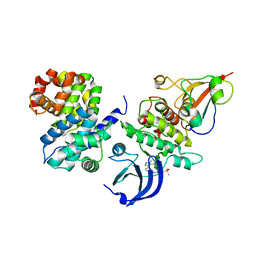 | | CDK1/CyclinB/Cks2 in complex with AZD5438 | | Descriptor: | 4-(2-methyl-3-propan-2-yl-imidazol-4-yl)-~{N}-(4-methylsulfonylphenyl)pyrimidin-2-amine, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, ... | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GU7
 
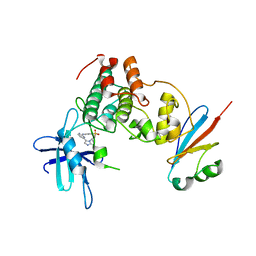 | | CDK1/Cks2 in complex with AZD5438 | | Descriptor: | 4-(2-methyl-3-propan-2-yl-imidazol-4-yl)-~{N}-(4-methylsulfonylphenyl)pyrimidin-2-amine, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
6GUF
 
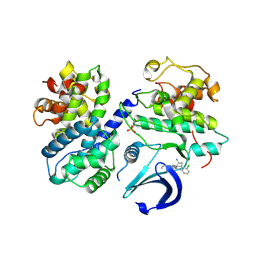 | | CDK2/CyclinA in complex with CGP74514A | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2, N2-[(1R,2S)-2-AMINOCYCLOHEXYL]-N6-(3-CHLOROPHENYL)-9-ETHYL-9H-PURINE-2,6-DIAMINE | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
7NGJ
 
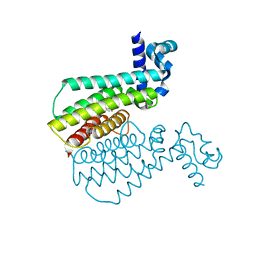 | |
7NGD
 
 | |
7NGO
 
 | |
7NGX
 
 | |
7NGK
 
 | |
7NGT
 
 | |
7NGU
 
 | |
7NGN
 
 | |
7NGR
 
 | |
7NGS
 
 | |
