2RNL
 
 | | Solution structure of the EGF-like domain from human Amphiregulin | | Descriptor: | Amphiregulin | | Authors: | Qin, X, Hayashi, F, Terada, T, Shirouzu, M, Watanabe, S, Kigawa, T, Yabuta, N, Nojima, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-01-11 | | Release date: | 2009-01-20 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the EGF-like domain from human Amphiregulin
To be Published
|
|
1S2R
 
 | | A High Resolution Crystal Structure of [d(CGCAAATTTGCG)]2 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3', SPERMINE | | Authors: | Woods, K.K, Maehigashi, T, Howerton, S.B, Tannenbaum, S, Williams, L.D. | | Deposit date: | 2004-01-09 | | Release date: | 2005-01-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | High-resolution structure of an extended A-tract: [d(CGCAAATTTGCG)]2.
J.Am.Chem.Soc., 126, 2004
|
|
6NIY
 
 | | A high-resolution cryo-electron microscopy structure of a calcitonin receptor-heterotrimeric Gs protein complex | | Descriptor: | Calcitonin, Calcitonin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | dal Maso, E, Glukhova, A, Zhu, Y, Garcia-Nafria, J, Tate, C.G, Atanasio, S, Reynolds, C.A, Ramirez-Aportela, E, Carazo, J.-M, Hick, C.A, Furness, S.G.B, Hay, D.L, Liang, Y.-L, Miller, L.J, Christopoulos, A, Wang, M.-W, Wootten, D, Sexton, P.M. | | Deposit date: | 2019-01-02 | | Release date: | 2019-01-23 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | The Molecular Control of Calcitonin Receptor Signaling.
Acs Pharmacol Transl Sci, 2, 2019
|
|
1BNA
 
 | | STRUCTURE OF A B-DNA DODECAMER. CONFORMATION AND DYNAMICS | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Drew, H.R, Wing, R.M, Takano, T, Broka, C, Tanaka, S, Itakura, K, Dickerson, R.E. | | Deposit date: | 1981-01-26 | | Release date: | 1981-05-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a B-DNA dodecamer: conformation and dynamics.
Proc.Natl.Acad.Sci.USA, 78, 1981
|
|
7OEX
 
 | |
8J7X
 
 | | Cryo-EM structure of hZnT7DeltaHis-loop-Fab complex in zinc-unbound state, determined in outward-facing conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, Zinc transporter 7 | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J7V
 
 | | Cryo-EM structure of hZnT7-Fab complex in zinc-unbound state, determined in heterogeneous conformations- one subunit in an inward-facing and the other in an outward-facing conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, Zinc transporter 7 | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J80
 
 | | Cryo-EM structure of hZnT7-Fab complex in zinc state 1, determined in heterogeneous conformations- one subunit in an inward-facing zinc-bound and the other in an outward-facing zinc-unbound conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, ZINC ION, ... | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J7U
 
 | | Cryo-EM structure of hZnT7-Fab complex in zinc-bound state, determined in outward-facing conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, ZINC ION, ... | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J7W
 
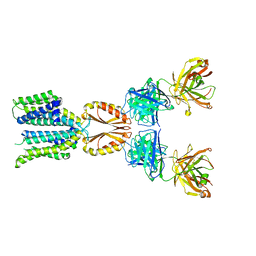 | | Cryo-EM structure of hZnT7-Fab complex in zinc state 2, determined in heterogeneous conformations- one subunit in an inward-facing zinc-bound and the other in an outward-facing zinc-bound conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, ZINC ION, ... | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J7T
 
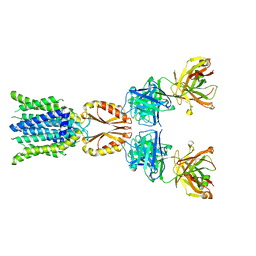 | | Cryo-EM structure of hZnT7-Fab complex in zinc-unbound state, determined in outward-facing conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, Zinc transporter 7 | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
8J7Y
 
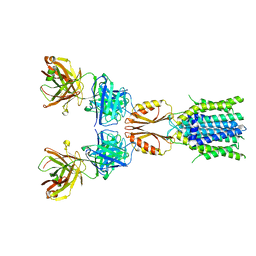 | | Cryo-EM structure of hZnT7DeltaHis-loop-Fab complex in zinc-bound state, determined in outward-facing conformation | | Descriptor: | Heavy chain of YN7114-08 Fab, Light chain of YN7114-08 Fab, ZINC ION, ... | | Authors: | Han, B.B, Inaba, K, Watanabe, S. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Nat Commun, 14, 2023
|
|
7LTO
 
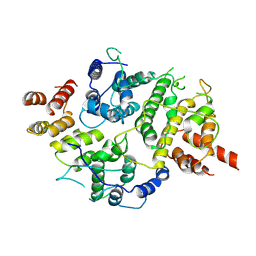 | | Nse5-6 complex | | Descriptor: | Non-structural maintenance of chromosome element 5, Ubiquitin-like protein SMT3,DNA repair protein KRE29 chimera | | Authors: | Yu, Y, Li, S.B, Zheng, S, Tangy, S, Koyi, C, Wan, B.B, Kung, H.H, Andrej, S, Alex, K, Patel, D.J, Zhao, X.L. | | Deposit date: | 2021-02-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Integrative analysis reveals unique structural and functional features of the Smc5/6 complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1NND
 
 | | Arginine 116 is Essential for Nucleic Acid Recognition by the Fingers Domain of Moloney Murine Leukemia Virus Reverse Transcriptase | | Descriptor: | Reverse Transcriptase | | Authors: | Crowther, R.L, Remeta, D.P, Minetti, C.A, Das, D, Montano, S.P, Georgiadis, M.M. | | Deposit date: | 2003-01-13 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and energetic characterization of nucleic acid-binding to the fingers domain of Moloney murine leukemia virus reverse transcriptase
Proteins, 57, 2004
|
|
4GCC
 
 | |
4GCD
 
 | |
3AM2
 
 | | Clostridium perfringens enterotoxin | | Descriptor: | GLYCEROL, Heat-labile enterotoxin B chain, UNKNOWN ATOM OR ION | | Authors: | Kitadokoro, K, Nishimura, K, Kamitani, S, Kimura, J, Fukui, A, Abe, H, Horiguchi, Y. | | Deposit date: | 2010-08-12 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of Clostridium perfringens Enterotoxin Displays Features of {beta}-Pore-forming Toxins
J.Biol.Chem., 286, 2011
|
|
3B4N
 
 | | Crystal Structure Analysis of Pectate Lyase PelI from Erwinia chrysanthemi | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Endo-pectate lyase, ... | | Authors: | Creze, C, Castang, S, Derivery, E, Haser, R, Shevchik, V, Gouet, P. | | Deposit date: | 2007-10-24 | | Release date: | 2008-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Crystal Structure of Pectate Lyase PelI from Soft Rot Pathogen Erwinia chrysanthemi in Complex with Its Substrate.
J.Biol.Chem., 283, 2008
|
|
2LEX
 
 | | Complex of the C-terminal WRKY domain of AtWRKY4 and a W-box DNA | | Descriptor: | DNA (5'-D(*CP*G*CP*CP*TP*TP*TP*GP*AP*CP*CP*AP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*C*TP*GP*GP*TP*CP*AP*AP*AP*GP*GP*CP*G)-3'), Probable WRKY transcription factor 4, ... | | Authors: | Yamasaki, K, Kigawa, T, Watanabe, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-06-24 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for sequence-spscific DNA recognition by an Arabidopsis WRKY transcription factor
J.Biol.Chem., 2012
|
|
6UD0
 
 | | Solution-state NMR structural ensemble of human Tsg101 UEV in complex with K63-linked diubiquitin | | Descriptor: | Tumor susceptibility gene 101 protein, Ubiquitin | | Authors: | Strickland, M, Watanabe, S, Bonn, S.M, Camara, C.M, Fushman, D, Carter, C.A, Tjandra, N. | | Deposit date: | 2019-09-18 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Tsg101/ESCRT-I Recruitment Regulated by the Dual Binding Modes of K63-Linked Diubiquitin
Structure, 2021
|
|
2LC2
 
 | | Solution structure of the RXLR effector P. capsici AVR3a4 | | Descriptor: | AVR3a4 | | Authors: | Li, H, Koshiba, S, Yaeno, T, Sato, M, Watanabe, S, Harada, T, Shirasu, K, Kigawa, T. | | Deposit date: | 2011-04-12 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A PIP-binding interface in the oomycete RXLR effector AVR3A is required for its accumulation in host cells to modulate plant immunity
To be Published
|
|
7XVL
 
 | | Crystal Structure of Nucleosome-H1.0 Linker Histone Assembly (sticky-169an DNA fragment) | | Descriptor: | DNA (169-MER), Histone H1.0, Histone H2A type 1-B/E, ... | | Authors: | Adhireksan, Z, Qiuye, B, Lee, P.L, Sharma, D, Padavattan, S, Davey, C.A. | | Deposit date: | 2022-05-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.506 Å) | | Cite: | Crystal Structure of Nucleosome-H1.0 Linker Histone Assembly (sticky-169an DNA fragment)
To Be Published
|
|
1IF2
 
 | | X-RAY STRUCTURE OF LEISHMANIA MEXICANA TRIOSEPHOSPHATE ISOMERASE COMPLEXED WITH IPP | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE, [2(FORMYL-HYDROXY-AMINO)-ETHYL]-PHOSPHONIC ACID | | Authors: | Kursula, I, Partanen, S, Lambeir, A.-M, Antonov, D.M, Augustyns, K, Wierenga, R.K. | | Deposit date: | 2001-04-12 | | Release date: | 2001-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural determinants for ligand binding and catalysis of triosephosphate isomerase.
Eur.J.Biochem., 268, 2001
|
|
8XWK
 
 | |
6UZT
 
 | | Crystal Structure of RPTP alpha | | Descriptor: | Receptor-type tyrosine-protein phosphatase alpha | | Authors: | Santelli, E, Wen, Y, Yang, S, Svensson, M.N.D, Stanford, S.M, Bottini, N. | | Deposit date: | 2019-11-15 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RPTP alpha phosphatase activity is allosterically regulated by the membrane-distal catalytic domain.
J.Biol.Chem., 295, 2020
|
|
