1NPE
 
 | | Crystal structure of Nidogen/Laminin Complex | | Descriptor: | CADMIUM ION, Laminin gamma-1 chain, nidogen | | Authors: | Takagi, J, Yang, Y.T, Liu, J.-H, Wang, J.-H, Springer, T.A. | | Deposit date: | 2003-01-17 | | Release date: | 2003-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complex between nidogen and laminin fragments reveals a paradigmatic
beta-propeller interface
Nature, 424, 2003
|
|
3GHN
 
 | | Crystal structure of the exosite-containing fragment of human ADAMTS13 (form-2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A disintegrin and metalloproteinase with thrombospondin motifs 13, ... | | Authors: | Akiyama, M, Takeda, S, Kokame, K, Takagi, J, Miyata, T. | | Deposit date: | 2009-03-04 | | Release date: | 2009-10-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the non-catalytic domains of ADAMTS13 reveal multiple discontinuous exosites for von Willebrand factor
To be Published
|
|
3GHM
 
 | | Crystal structure of the exosite-containing fragment of human ADAMTS13 (form-1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A disintegrin and metalloproteinase with thrombospondin motifs 13, ... | | Authors: | Akiyama, M, Takeda, S, Kokame, K, Takagi, J, Miyata, T. | | Deposit date: | 2009-03-04 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the non-catalytic domains of ADAMTS13 reveal multiple discontinuous exosites for von Willebrand factor
To be Published
|
|
5GJE
 
 | | Three-dimensional reconstruction of human LRP6 ectodomain complexed with Dkk1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Matoba, K, Mihara, E, Tamura-Kawakami, K, Hirai, H, Thompson, S, Iwasaki, K, Takagi, J. | | Deposit date: | 2016-06-29 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (21 Å) | | Cite: | Conformational Freedom of the LRP6 Ectodomain Is Regulated by N-glycosylation and the Binding of the Wnt Antagonist Dkk1
Cell Rep, 18, 2017
|
|
7XOM
 
 | |
7XOK
 
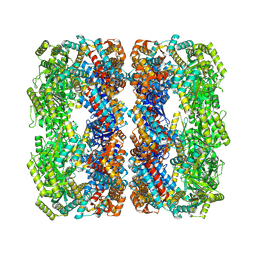 | |
5GKR
 
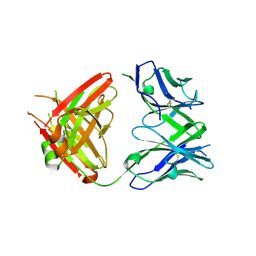 | | Crystal structure of SLE patient-derived anti-DNA antibody in complex with oligonucleotide | | Descriptor: | DNA (5'-D(P*TP*TP*TP*T)-3'), IgG2, Fab (heavy chain), ... | | Authors: | Arimori, T, Sakakibara, S, Kikutani, H, Takagi, J. | | Deposit date: | 2016-07-05 | | Release date: | 2017-07-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Clonal evolution and antigen recognition of anti-nuclear antibodies in acute systemic lupus erythematosus
Sci Rep, 7, 2017
|
|
5GKS
 
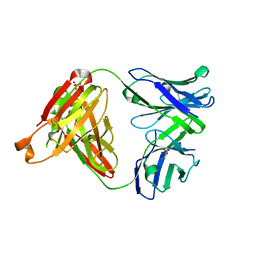 | | Crystal structure of SLE patient-derived anti-DNA antibody | | Descriptor: | IgG2, Fab (heavy chain), PHOSPHATE ION, ... | | Authors: | Arimori, T, Sakakibara, S, Kikutani, H, Takagi, J. | | Deposit date: | 2016-07-05 | | Release date: | 2017-07-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Clonal evolution and antigen recognition of anti-nuclear antibodies in acute systemic lupus erythematosus
Sci Rep, 7, 2017
|
|
4YO0
 
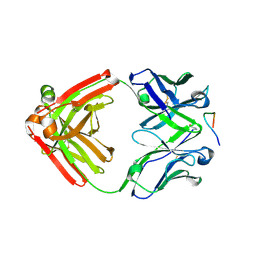 | | Crystal structure of monoclonal anti-human podoplanin antibody NZ-1 with bound PA peptide | | Descriptor: | 1,2-ETHANEDIOL, Heavy chain of antigen binding fragment, Fab, ... | | Authors: | Fujii, Y, Kitago, Y, Arimori, T, Takagi, J. | | Deposit date: | 2015-03-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Tailored placement of a turn-forming PA tag into the structured domain of a protein to probe its conformational state
J.Cell.Sci., 129, 2016
|
|
5GIS
 
 | | Crystal structure of a Fab fragment with its ligand peptide | | Descriptor: | Heavy chain of Fab fragment, Light chain of Fab fragment, SULFATE ION, ... | | Authors: | Kitago, Y, Kaneko, K.K, Ogasawara, S, Kato, Y, Takagi, J. | | Deposit date: | 2016-06-24 | | Release date: | 2016-09-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis for multi-specific peptide recognition by the anti-IDH1/2 monoclonal antibody, MsMab-1.
Biochem. Biophys. Res. Commun., 478, 2016
|
|
1MQA
 
 | | Crystal structure of high affinity alphaL I domain in the absence of ligand or metal | | Descriptor: | Integrin alpha-L | | Authors: | Shimaoka, T, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, Zhang, R, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
1MQ9
 
 | | Crystal structure of high affinity alphaL I domain with ligand mimetic crystal contact | | Descriptor: | Integrin alpha-L, MANGANESE (II) ION | | Authors: | Shimaoka, M, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, McCormack, A, Zhang, R, Joachimiak, A, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
5GIR
 
 | | Crystal structure of a Fab fragment with its ligand peptide | | Descriptor: | 1,2-ETHANEDIOL, Heavy chain of Fab fragment, LYS-PRO-ILE-ILE-ILE-GLY-SER-HIS-ALA-TYR-GLY-ASP, ... | | Authors: | Kitago, Y, Kaneko, K.K, Ogasawara, S, Kato, Y, Takagi, J. | | Deposit date: | 2016-06-24 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis for multi-specific peptide recognition by the anti-IDH1/2 monoclonal antibody, MsMab-1.
Biochem. Biophys. Res. Commun., 478, 2016
|
|
7NWL
 
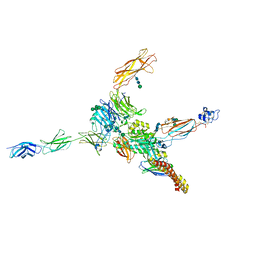 | | Cryo-EM structure of human integrin alpha5beta1 (open form) in complex with fibronectin and TS2/16 Fv-clasp | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Integrin alpha-5, ... | | Authors: | Schumacher, S, Dedden, D, Vazquez Nunez, R, Matoba, K, Takagi, J, Biertumpfel, C, Mizuno, N. | | Deposit date: | 2021-03-17 | | Release date: | 2021-06-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into integrin alpha 5 beta 1 opening by fibronectin ligand.
Sci Adv, 7, 2021
|
|
7NXD
 
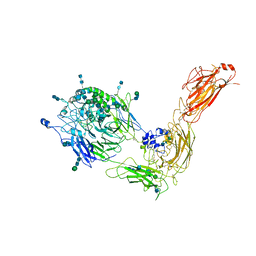 | | Cryo-EM structure of human integrin alpha5beta1 in the half-bent conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Schumacher, S, Dedden, D, Vazquez Nunez, R, Matoba, K, Takagi, J, Biertumpfel, C, Mizuno, N. | | Deposit date: | 2021-03-18 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural insights into integrin alpha 5 beta 1 opening by fibronectin ligand.
Sci Adv, 7, 2021
|
|
5XCU
 
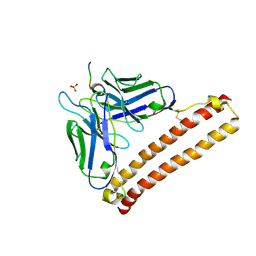 | | Crystal structure of 12CA5 Fv-clasp fragment with its antigen peptide | | Descriptor: | HA peptide, SULFATE ION, VH(S112C)-SARAH chimera,VH(S112C)-SARAH chimera, ... | | Authors: | Arimori, T, Takagi, J. | | Deposit date: | 2017-03-23 | | Release date: | 2017-10-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.454 Å) | | Cite: | Fv-clasp: An Artificially Designed Small Antibody Fragment with Improved Production Compatibility, Stability, and Crystallizability
Structure, 25, 2017
|
|
5XCQ
 
 | | Crystal structure of P20.1 Fv-clasp fragment with its antigen peptide | | Descriptor: | C8 peptide, SULFATE ION, VH-SARAH(Y35C)chimera, ... | | Authors: | Arimori, T, Takagi, J. | | Deposit date: | 2017-03-23 | | Release date: | 2017-10-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.313 Å) | | Cite: | Fv-clasp: An Artificially Designed Small Antibody Fragment with Improved Production Compatibility, Stability, and Crystallizability
Structure, 25, 2017
|
|
5XCV
 
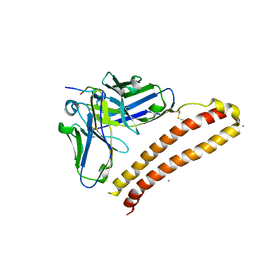 | | Crystal structure of NZ-1 Fv-clasp fragment with its antigen peptide | | Descriptor: | CALCIUM ION, PA14 peptide, VH(S112C)-SARAH chimera,VH(S112C)-SARAH chimera, ... | | Authors: | Arimori, T, Takagi, J. | | Deposit date: | 2017-03-23 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.143 Å) | | Cite: | Fv-clasp: An Artificially Designed Small Antibody Fragment with Improved Production Compatibility, Stability, and Crystallizability
Structure, 25, 2017
|
|
5XCT
 
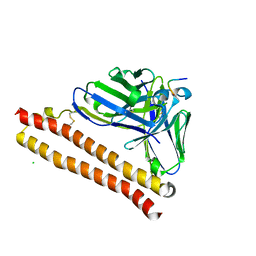 | | Crystal structure of P20.1 Fv-clasp fragment with its antigen peptide | | Descriptor: | C8 peptide, CHLORIDE ION, VH(S112C)-SARAH chimera, ... | | Authors: | Arimori, T, Takagi, J. | | Deposit date: | 2017-03-23 | | Release date: | 2017-10-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Fv-clasp: An Artificially Designed Small Antibody Fragment with Improved Production Compatibility, Stability, and Crystallizability
Structure, 25, 2017
|
|
7VF3
 
 | | Plexin B1 extracellular fragment in complex with lasso-grafted PB1m7 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Plexin-B1, ... | | Authors: | Sugano, N.N, Hirata, K, Yamashita, K, Yamamoto, M, Arimori, T, Takagi, J. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | De novo Fc-based receptor dimerizers differentially modulate PlexinB1 function.
Structure, 30, 2022
|
|
8JYR
 
 | |
8JYQ
 
 | |
7DMU
 
 | |
7XOJ
 
 | |
7XON
 
 | |
