5HQ7
 
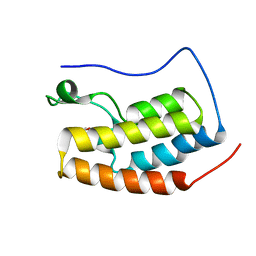 | | Crystal structure of fragment bound with Brd4 | | Descriptor: | Bromodomain-containing protein 4, N-ethyl-6,7-dimethoxyquinazolin-4-amine | | Authors: | Chen, T.T, Xu, Y.C. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of fragment bound with Brd4
to be published
|
|
4GG7
 
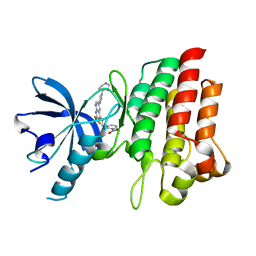 | | Crystal structure of cMET in complex with novel inhibitor | | Descriptor: | Hepatocyte growth factor receptor, N-(3-nitrobenzyl)-6-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]-2-(trifluoromethyl)pyrido[2,3-d]pyrimidin-4-amine | | Authors: | Liu, Q.F, Chen, T.T, Xu, Y.C. | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Multisubstituted quinoxalines and pyrido[2,3-d]pyrimidines: Synthesis and SAR study as tyrosine kinase c-Met inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1II8
 
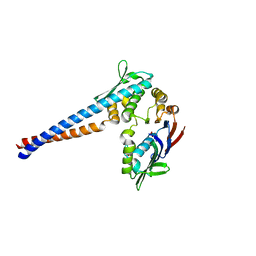 | | Crystal structure of the P. furiosus Rad50 ATPase domain | | Descriptor: | PHOSPHATE ION, Rad50 ABC-ATPase | | Authors: | Hopfner, K.-P, Karcher, A, Craig, L, Woo, T.T, Carney, J.P, Tainer, J.A. | | Deposit date: | 2001-04-20 | | Release date: | 2001-05-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase.
Cell(Cambridge,Mass.), 105, 2001
|
|
4GEV
 
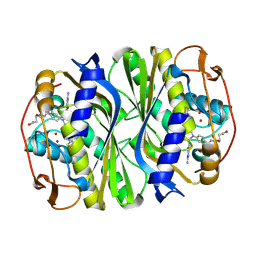 | | E. coli thymidylate synthase Y209W variant in complex with substrate and a cofactor analog | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-deoxy-5'-uridylic acid, Thymidylate synthase | | Authors: | Newby, Z, Lee, T.T, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2012-08-02 | | Release date: | 2012-08-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A remote mutation affects the hydride transfer by disrupting concerted protein motions in thymidylate synthase.
J.Am.Chem.Soc., 134, 2012
|
|
1II7
 
 | | Crystal structure of P. furiosus Mre11 with manganese and dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, Mre11 nuclease, ... | | Authors: | Hopfner, K.-P, Karcher, A, Craig, L, Woo, T.T, Carney, J.P, Tainer, J.A. | | Deposit date: | 2001-04-20 | | Release date: | 2001-05-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase.
Cell(Cambridge,Mass.), 105, 2001
|
|
7S94
 
 | |
7SBN
 
 | |
7SBM
 
 | | Human glutaminase C (Y466W) with L-Gln, open conformation | | Descriptor: | GLUTAMINE, Isoform 3 of Glutaminase kidney isoform, mitochondrial | | Authors: | Nguyen, T.-T.T, Cerione, R.A. | | Deposit date: | 2021-09-25 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High-resolution structures of mitochondrial glutaminase C tetramers indicate conformational changes upon phosphate binding.
J.Biol.Chem., 298, 2022
|
|
5KE4
 
 | | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) containing loop C from the human alpha 6 nicotinic acetylcholine receptor in complex with 2-((5-(3,7-Diazabicyclo[3.3.1]nonan-3-yl)pyridin-3-yl)oxy)- N,N-dimethylethanamine (BPC) | | Descriptor: | 2-((5-(3,7-Diazabicyclo[3.3.1]nonan-3-yl)pyridin-3-yl)oxy)-N,N-dimethylethanamine, Soluble acetylcholine receptor | | Authors: | Bobango, J, Wu, J, Talley, I.T, Ralston, R, Sankaran, B, Talley, T.T. | | Deposit date: | 2016-06-09 | | Release date: | 2016-07-06 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) containing loop C from the human alpha 6 nicotinic acetylcholine receptor in complex with 2-((5-(3,7-Diazabicyclo[3.3.1]nonan-3-yl)pyridin-3-yl)oxy)-
N,N-dimethylethanamine (BPC)
To Be Published
|
|
5JME
 
 | |
8T0Z
 
 | | Human liver-type glutaminase (K253A) with L-Gln, filamentous form | | Descriptor: | GLUTAMINE, Glutaminase liver isoform, mitochondrial | | Authors: | Feng, S, Aplin, C, Nguyen, T.-T.T, Milano, S.K, Cerione, R.A. | | Deposit date: | 2023-06-01 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Filament formation drives catalysis by glutaminase enzymes important in cancer progression.
Nat Commun, 15, 2024
|
|
8SZL
 
 | | Human liver-type glutaminase (Apo form) | | Descriptor: | Glutaminase liver isoform, mitochondrial | | Authors: | Feng, S, Aplin, C, Nguyen, T.-T.T, Milano, S.K, Cerione, R.A. | | Deposit date: | 2023-05-30 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Filament formation drives catalysis by glutaminase enzymes important in cancer progression.
Nat Commun, 15, 2024
|
|
8SZJ
 
 | | Human glutaminase C (Y466W) with L-Gln and Pi, filamentous form | | Descriptor: | GLUTAMINE, Glutaminase kidney isoform, mitochondrial, ... | | Authors: | Feng, S, Aplin, C, Nguyen, T.-T.T, Milano, S.K, Cerione, R.A. | | Deposit date: | 2023-05-29 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Filament formation drives catalysis by glutaminase enzymes important in cancer progression.
Nat Commun, 15, 2024
|
|
5KZU
 
 | | Crystal structure of an acetylcholine binding protein from Aplysia californica (Ac-AChBP) in complex with click chemistry compound 9-[[1-[8-methyl-8-(2-phenylethyl)-8-azoniabicyclo[3.2.1]octan-3-yl]triazol-4-yl]methyl]carbazole | | Descriptor: | 9-[[1-[8-methyl-8-(2-phenylethyl)-8-azoniabicyclo[3.2.1]octan-3-yl]triazol-4-yl]methyl]carbazole, SULFATE ION, Soluble acetylcholine receptor, ... | | Authors: | Bobango, J, Wu, J, Talley, I.T, Sankaran, B, Talley, T.T. | | Deposit date: | 2016-07-25 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) in complex with click chemistry compound 9-[[1-[8-methyl-8-(2-phenylethyl)-8-azoniabicyclo[3.2.1]octan-3-yl]triazol-4-yl]methyl]carbazole
To Be Published
|
|
3C84
 
 | | Crystal structure of a complex of AChBP from aplysia californica and the neonicotinoid thiacloprid | | Descriptor: | ISOPROPYL ALCOHOL, MAGNESIUM ION, Soluble acetylcholine receptor, ... | | Authors: | Talley, T.T, Harel, M, Hibbs, R.E, Tomizawa, M, Casida, J.E, Taylor, P.W. | | Deposit date: | 2008-02-08 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Atomic interactions of neonicotinoid agonists with AChBP: molecular recognition of the distinctive electronegative pharmacophore.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BG4
 
 | | The crystal structure of guamerin in complex with chymotrypsin and the development of an elastase-specific inhibitor | | Descriptor: | Chymotrypsin A chain A, Chymotrypsin A chain B, Chymotrypsin A chain C, ... | | Authors: | Kim, H, Chu, T.T.T, Kim, D.Y, Kim, D.R, Nguyen, C.M.T, Choi, J, Lee, J.R, Hahn, M.J, Kim, K.K. | | Deposit date: | 2007-11-26 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of guamerin in complex with chymotrypsin and the development of an elastase-specific inhibitor.
J.Mol.Biol., 376, 2008
|
|
3C79
 
 | | Crystal structure of Aplysia californica AChBP in complex with the neonicotinoid imidacloprid | | Descriptor: | (2E)-1-[(6-chloropyridin-3-yl)methyl]-N-nitroimidazolidin-2-imine, ISOPROPYL ALCOHOL, Soluble acetylcholine receptor | | Authors: | Talley, T.T, Harel, M, Hibbs, R.E, Tomizawa, M, Casida, J.E, Taylor, P.W. | | Deposit date: | 2008-02-06 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Atomic interactions of neonicotinoid agonists with AChBP: molecular recognition of the distinctive electronegative pharmacophore.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
8ZRK
 
 | | Cryo-EM structure of GPR119-Gs Complex with small molecule agonist GSK-1292263 | | Descriptor: | 5-[4-[[6-(4-methylsulfonylphenyl)pyridin-3-yl]oxymethyl]piperidin-1-yl]-3-propan-2-yl-1,2,4-oxadiazole, Glucose-dependent insulinotropic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wong, T.S, Xiong, T.T, Zeng, Z.C, Gan, S.Y, Qiu, C, Du, Y. | | Deposit date: | 2024-06-04 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Cryo-EM structure of GPR119-Gs complex
To be published
|
|
8UTG
 
 | | An RNA G-Quadruplex from the NS5 gene in the West Nile Virus Genome | | Descriptor: | AMMONIUM ION, POTASSIUM ION, RNA (5'-R(*UP*AP*UP*GP*GP*AP*AP*GP*AP*GP*GP*CP*GP*GP*UP*UP*GP*GP*UP*AP*U)-3') | | Authors: | Terrell, J.R, Le, T.T, Wilson, W.D, Siemer, J.L. | | Deposit date: | 2023-10-31 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of an RNA G-quadruplex from the West Nile virus genome.
Nat Commun, 15, 2024
|
|
8ZR5
 
 | | Cryo-EM Structure of GPR119-Gs-Firuglipel complex | | Descriptor: | 4-[5-[(1~{R})-1-(4-cyclopropylcarbonylphenoxy)propyl]-1,2,4-oxadiazol-3-yl]-2-fluoranyl-~{N}-[(2~{R})-1-oxidanylpropan-2-yl]benzamide, Glucose-dependent insulinotropic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wong, T.S, Zeng, Z.C, Xiong, T.T, Gan, S.Y, Qiu, C, Du, Y. | | Deposit date: | 2024-06-04 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Cryo-EM structure of GPR119-Gs complex
To Be Published
|
|
7S55
 
 | |
1OI0
 
 | | CRYSTAL STRUCTURE OF AF2198, A JAB1/MPN DOMAIN PROTEIN FROM ARCHAEOGLOBUS FULGIDUS | | Descriptor: | HYPOTHETICAL PROTEIN AF2198, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM, ZINC ION | | Authors: | Tran, H.J.T.T, Allen, M.D, Lowe, J, Bycroft, M. | | Deposit date: | 2003-06-04 | | Release date: | 2003-08-14 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of the Jab1/Mpn Domain and its Implications for Proteasome Function
Biochemistry, 42, 2003
|
|
4V3N
 
 | | Membrane bound pleurotolysin prepore (TMH2 strand lock) trapped with engineered disulphide cross-link | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-20 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
4V3A
 
 | | Membrane bound pleurotolysin prepore (TMH1 lock) trapped with engineered disulphide cross-link | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, CaradocDavies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-17 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
4V3M
 
 | | Membrane bound pleurotolysin prepore (TMH2 helix lock) trapped with engineered disulphide cross-link | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-20 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
