4YQV
 
 | | Glutathione S-transferase Omega 1 bound to covalent inhibitor C4-10 | | Descriptor: | 2-(ethylsulfanyl)-N-methyl-N-[(1-phenyl-1H-pyrazol-4-yl)methyl]acetamide, Glutathione S-transferase omega-1 | | Authors: | Stuckey, J.A. | | Deposit date: | 2015-03-13 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Mechanistic evaluation and transcriptional signature of a glutathione S-transferase omega 1 inhibitor.
Nat Commun, 7, 2016
|
|
4YQU
 
 | | Glutathione S-transferase Omega 1 bound to covalent inhibitor C1-31 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Glutathione S-transferase omega-1, N-{5-(azepan-1-ylsulfonyl)-2-[(ethylsulfanyl)methoxy]phenyl}acetamide | | Authors: | Stuckey, J.A. | | Deposit date: | 2015-03-13 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanistic evaluation and transcriptional signature of a glutathione S-transferase omega 1 inhibitor.
Nat Commun, 7, 2016
|
|
9BIG
 
 | | Stat6 bound to degrader AK-1690 | | Descriptor: | Signal transducer and activator of transcription 6, [(2-{[(2S)-1-{(2S,4S)-4-[(7-{2-[(3R)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}hept-6-yn-1-yl)oxy]-2-[(2R)-2-phenylmorpholine-4-carbonyl]pyrrolidin-1-yl}-3,3-dimethyl-1-oxobutan-2-yl]carbamoyl}-1-benzothiophen-5-yl)di(fluoro)methyl]phosphonic acid | | Authors: | Mallik, L, Stuckey, J.A. | | Deposit date: | 2024-04-23 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (3.304 Å) | | Cite: | Discovery of AK-1690: A Potent and Highly Selective STAT6 PROTAC Degrader.
J.Med.Chem., 2024
|
|
8GBL
 
 | | Structure of Apo Human SIRT5 | | Descriptor: | NAD-dependent protein deacylase sirtuin-5, mitochondrial, ZINC ION | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2023-02-26 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Human SIRT5 variants with reduced stability and activity do not cause neuropathology in mice.
Iscience, 27, 2024
|
|
8GBN
 
 | | Structure of Apo Human SIRT5 P114T Mutant | | Descriptor: | 1,2-ETHANEDIOL, NAD-dependent protein deacylase sirtuin-5, mitochondrial, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2023-02-26 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human SIRT5 variants with reduced stability and activity do not cause neuropathology in mice.
Iscience, 27, 2024
|
|
8SX4
 
 | | Crystal Structure of eIF4e in complex with Compound 7n | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Eukaryotic translation initiation factor 4E, [(~{Z})-4-[2-azanyl-7-[(5-chloranyl-1~{H}-indol-2-yl)methyl]-6-oxidanylidene-1~{H}-purin-9-yl]but-2-enyl]phosphonic acid | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2023-05-19 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Design of Cell-Permeable Inhibitors of Eukaryotic Translation Initiation Factor 4E (eIF4E) for Inhibiting Aberrant Cap-Dependent Translation in Cancer.
J.Med.Chem., 66, 2023
|
|
6EGP
 
 | |
6EGN
 
 | | Crystal Structure of a Three-stranded Coiled Coil Peptide Containing a Trigonal Planar Hg(II)S3 Site Modified by D-Leu in the Second Coordination Sphere | | Descriptor: | CHLORIDE ION, Hg(II)(GRAND CoilSerL16CL19(DLE))3-, MERCURY (II) ION, ... | | Authors: | Ruckthong, L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2018-08-20 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | How Outer Coordination Sphere Modifications Can Impact Metal Structures in Proteins: A Crystallographic Evaluation.
Chemistry, 25, 2019
|
|
6EGO
 
 | |
6EGM
 
 | |
6EGL
 
 | |
4TW3
 
 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Bound | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde decarbonylase, FE (III) ION, ... | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-06-29 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
6U6F
 
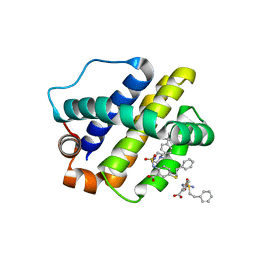 | | The crystal structure of anti-apoptotic Mcl-1 protein in complex with 2, 5-substituted benzoic acid inhibitor 21 | | Descriptor: | 2-[({4-[(4-tert-butylphenyl)methyl]piperazin-1-yl}sulfonyl)amino]-5-[(2-phenylethyl)sulfanyl]benzoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Yang, Y, Stuckey, J.A, Nikolovska-Coleska, Z. | | Deposit date: | 2019-08-29 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery and Characterization of 2,5-Substituted Benzoic Acid Dual Inhibitors of the Anti-apoptotic Mcl-1 and Bfl-1 Proteins.
J.Med.Chem., 63, 2020
|
|
6U9K
 
 | | MLL1 SET N3861I/Q3867L bound to inhibitor 18 (TC-5153) | | Descriptor: | 5'-([(3S)-3-amino-3-carboxypropyl]{[1-(3,3-diphenylpropyl)azetidin-3-yl]methyl}amino)-5'-deoxyadenosine, GLYCEROL, Histone-lysine N-methyltransferase, ... | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of MLL Methyltransferase.
Acs Med.Chem.Lett., 11, 2020
|
|
6U9N
 
 | | MLL1 SET N3861I/Q3867L bound to inhibitor 14 (TC-5139) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl]({1-[(4-chlorophenyl)methyl]azetidin-3-yl}methyl)amino}-5'-deoxyadenosine, Histone-lysine N-methyltransferase, ZINC ION | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of MLL Methyltransferase.
Acs Med.Chem.Lett., 11, 2020
|
|
6U9R
 
 | | MLL1 SET N3861I/Q3867L bound to inhibitor 12 (TC-5140) | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl]({1-[(3-chlorophenyl)methyl]azetidin-3-yl}methyl)amino}-5'-deoxyadenosine, Histone-lysine N-methyltransferase, ZINC ION | | Authors: | Petrunak, E.M, Stuckey, J.A. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of MLL Methyltransferase.
Acs Med.Chem.Lett., 11, 2020
|
|
3PBJ
 
 | | Hydrolytic catalysis and structural stabilization in a designed metalloprotein | | Descriptor: | CHLORIDE ION, COIL SER L9L-Pen L23H, MERCURY (II) ION, ... | | Authors: | Zastrow, M.L, Peacock, A.F.A, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2010-10-20 | | Release date: | 2011-11-30 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hydrolytic catalysis and structural stabilization in a designed metalloprotein.
Nat Chem, 4, 2012
|
|
7TVA
 
 | | Stat5a Core in complex with AK2292 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MALONATE ION, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N-(5-{2-[(3R)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}pent-4-yn-1-yl)-N-methyl-N~3~-[4-(1,3-thiazol-2-yl)phenyl]-beta-alaninamide, ... | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-02-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.835 Å) | | Cite: | A selective small-molecule STAT5 PROTAC degrader capable of achieving tumor regression in vivo.
Nat.Chem.Biol., 19, 2023
|
|
7TVB
 
 | | Stat5A Core in Complex with AK305 | | Descriptor: | GLYCEROL, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N,N-dimethyl-N~3~-[4-(1,3-thiazol-2-yl)phenyl]-beta-alaninamide, Signal transducer and activator of transcription 5A | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-02-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | A selective small-molecule STAT5 PROTAC degrader capable of achieving tumor regression in vivo.
Nat.Chem.Biol., 19, 2023
|
|
7UC6
 
 | | Stat5a Core in complex with Compound 12 | | Descriptor: | N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N~3~-(4-bromophenyl)-N,N-dimethyl-beta-alaninamide, Signal transducer and activator of transcription 5A | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-03-16 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Discovery of a Potent and Selective STAT5 PROTAC Degrader with Strong Antitumor Activity In Vivo in Acute Myeloid Leukemia.
J.Med.Chem., 66, 2023
|
|
7UBT
 
 | | Stat5a Core in complex with Compound 18 | | Descriptor: | GLYCEROL, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N~3~-(1,3-benzothiazol-5-yl)-N,N-dimethyl-beta-alaninamide, Signal transducer and activator of transcription 5A | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-03-15 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Discovery of a Potent and Selective STAT5 PROTAC Degrader with Strong Antitumor Activity In Vivo in Acute Myeloid Leukemia.
J.Med.Chem., 66, 2023
|
|
7UC7
 
 | | Stat5a Core in complex with Compound 17 | | Descriptor: | GLYCEROL, N-{5-[difluoro(phosphono)methyl]-1-benzothiophene-2-carbonyl}-3-methyl-L-valyl-L-prolyl-N~3~-(1-benzofuran-5-yl)-N,N-dimethyl-beta-alaninamide, Signal transducer and activator of transcription 5A | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2022-03-16 | | Release date: | 2023-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Discovery of a Potent and Selective STAT5 PROTAC Degrader with Strong Antitumor Activity In Vivo in Acute Myeloid Leukemia.
J.Med.Chem., 66, 2023
|
|
2BN0
 
 | | Banana Lectin bound to Laminaribiose | | Descriptor: | CADMIUM ION, RIPENING-ASSOCIATED PROTEIN, SULFATE ION, ... | | Authors: | Meagher, J.L, Winter, H.C, Ezell, P, Goldstein, I.J, Stuckey, J.A. | | Deposit date: | 2005-03-17 | | Release date: | 2005-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Banana Lectin Reveals a Novel Second Sugar Binding Site.
Glycobiology, 15, 2005
|
|
2BMZ
 
 | | Banana Lectin bound to Xyl-b1,3 Man-a-O-Methyl (XM) | | Descriptor: | CADMIUM ION, RIPENING-ASSOCIATED PROTEIN, SULFATE ION, ... | | Authors: | Meagher, J.L, Winter, H.C, Ezell, P, Goldstein, I.J, Stuckey, J.A. | | Deposit date: | 2005-03-17 | | Release date: | 2005-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Banana Lectin Reveals a Novel Second Sugar Binding Site.
Glycobiology, 15, 2005
|
|
2BMY
 
 | | Banana Lectin | | Descriptor: | CADMIUM ION, RIPENING-ASSOCIATED PROTEIN, SULFATE ION | | Authors: | Meagher, J.L, Winter, H.C, Ezell, P, Goldstein, I.J, Stuckey, J.A. | | Deposit date: | 2005-03-17 | | Release date: | 2005-06-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Banana Lectin Reveals a Novel Second Sugar Binding Site.
Glycobiology, 15, 2005
|
|
