4XP8
 
 | |
4S2J
 
 | | OXA-48 in complex with Avibactam at pH 6.5 | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2015-01-20 | | Release date: | 2015-02-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Molecular Mechanism of Avibactam-Mediated beta-Lactamase Inhibition.
ACS Infect Dis, 1, 2015
|
|
8EXP
 
 | | Cryo-EM structure of S. aureus BlaR1 with C2 symmetry | | Descriptor: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | Authors: | Worrall, L.J, Alexander, J.A.N, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
8EXS
 
 | | Cryo-EM structure of S. aureus BlaR1 F284A mutant | | Descriptor: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | Authors: | Alexander, J.A.N, Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
8EXR
 
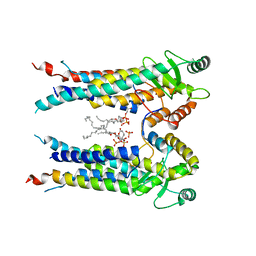 | | Cryo-EM structure of S. aureus BlaR1 TM and zinc metalloprotease domain | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Beta-lactam sensor/signal transducer BlaR1, PHOSPHATE ION, ... | | Authors: | Worrall, L.J, Alexander, J.A.N, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
8EXT
 
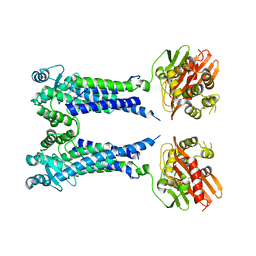 | | Cryo-EM structure of S. aureus BlaR1 F284A mutant in complex with ampicillin | | Descriptor: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | Authors: | Alexander, J.A.N, Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
4S2O
 
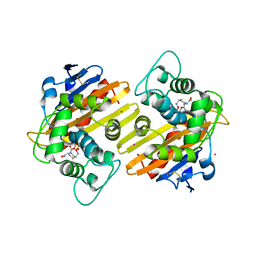 | | OXA-10 in complex with Avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase OXA-10, COBALT (II) ION | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2015-01-21 | | Release date: | 2015-02-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Mechanism of Avibactam-Mediated beta-Lactamase Inhibition.
ACS Infect Dis, 1, 2015
|
|
8EXQ
 
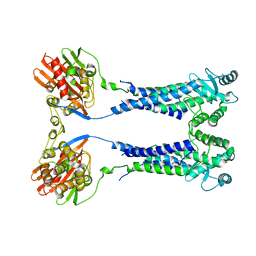 | | Cryo-EM structure of S. aureus BlaR1 with C1 symmetry | | Descriptor: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | Authors: | Worrall, L.J, Alexander, J.A.N, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
4S2N
 
 | | OXA-48 in complex with Avibactam at pH 8.5 | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2015-01-21 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Mechanism of Avibactam-Mediated beta-Lactamase Inhibition.
ACS Infect Dis, 1, 2015
|
|
4S2I
 
 | | CTX-M-15 in complex with Avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2015-01-20 | | Release date: | 2015-02-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Mechanism of Avibactam-Mediated beta-Lactamase Inhibition.
ACS Infect Dis, 1, 2015
|
|
4S2K
 
 | | OXA-48 in complex with Avibactam at pH 7.5 | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2015-01-20 | | Release date: | 2015-02-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Mechanism of Avibactam-Mediated beta-Lactamase Inhibition.
ACS Infect Dis, 1, 2015
|
|
4S2P
 
 | |
8FFJ
 
 | | Structure of Zanidatamab bound to HER2 | | Descriptor: | Receptor tyrosine-protein kinase erbB-2, Zanidatamab Heavy Chain A, Zanidatamab Heavy Chain B, ... | | Authors: | Worrall, L.J, Atkinson, C.E, Sanches, M, Dixit, S, Strynadka, N.C.J. | | Deposit date: | 2022-12-08 | | Release date: | 2023-02-22 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | An anti-HER2 biparatopic antibody that induces unique HER2 clustering and complement-dependent cytotoxicity.
Nat Commun, 14, 2023
|
|
4OYC
 
 | |
3T07
 
 | | Crystal structure of S. aureus Pyruvate Kinase in complex with a naturally occurring bis-indole alkaloid | | Descriptor: | (3S,5R)-3,5-bis(6-bromo-1H-indol-3-yl)piperazin-2-one, PHOSPHATE ION, Pyruvate kinase | | Authors: | Worrall, L.J, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2011-07-19 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Methicillin-resistant Staphylococcus aureus (MRSA) pyruvate kinase as a target for bis-indole alkaloids with antibacterial activities.
J.Biol.Chem., 286, 2011
|
|
3T05
 
 | |
4Q6V
 
 | |
4Q6Z
 
 | | LpoB C-terminal domain from Escherichia coli | | Descriptor: | Penicillin-binding protein activator LpoB | | Authors: | King, D.T, Strynadka, N.C.J. | | Deposit date: | 2014-04-23 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Lipoprotein Outer Membrane Regulator of Penicillin-binding Protein 1B.
J.Biol.Chem., 289, 2014
|
|
4Q6L
 
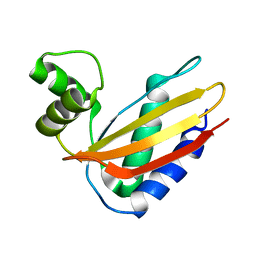 | |
1SN8
 
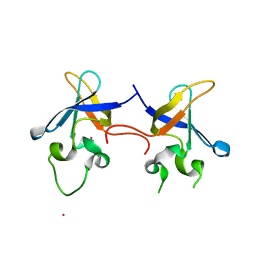 | | Crystal structure of the S1 domain of RNase E from E. coli (Pb derivative) | | Descriptor: | LEAD (II) ION, Ribonuclease E | | Authors: | Schubert, M, Edge, R.E, Lario, P, Cook, M.A, Strynadka, N.C.J, Mackie, G.A, McIntosh, L.P. | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
J.Mol.Biol., 341, 2004
|
|
1SMX
 
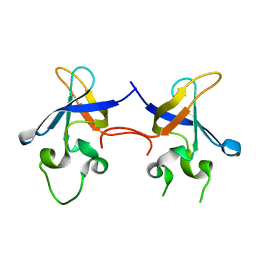 | | Crystal structure of the S1 domain of RNase E from E. coli (native) | | Descriptor: | Ribonuclease E | | Authors: | Schubert, M, Edge, R.E, Lario, P, Cook, M.A, Strynadka, N.C.J, Mackie, G.A, McIntosh, L.P. | | Deposit date: | 2004-03-09 | | Release date: | 2004-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
J.Mol.Biol., 341, 2004
|
|
6UEX
 
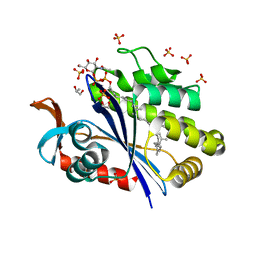 | | Crystal structure of S. aureus LcpA in complex with octaprenyl-pyrophosphate-GlcNAc | | Descriptor: | 2-(acetylamino)-2-deoxy-1-O-[(S)-hydroxy{[(S)-hydroxy{[(2Z,6Z,10Z,14Z,18Z,22Z,26Z)-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaen-1-yl]oxy}phosphoryl]oxy}phosphoryl]-alpha-D-glucopyranose, GLYCEROL, Regulatory protein MsrR, ... | | Authors: | Li, F.K.K, Strynadka, N.C.J. | | Deposit date: | 2019-09-23 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic analysis ofStaphylococcus aureusLcpA, the primary wall teichoic acid ligase.
J.Biol.Chem., 295, 2020
|
|
3L7L
 
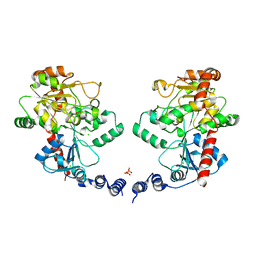 | | Structure of the Wall Teichoic Acid Polymerase TagF, H444N + CDPG (30 minute soak) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Lovering, A.L, Strynadka, N.C.J. | | Deposit date: | 2009-12-28 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the bacterial teichoic acid polymerase TagF provides insights into membrane association and catalysis.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6UF6
 
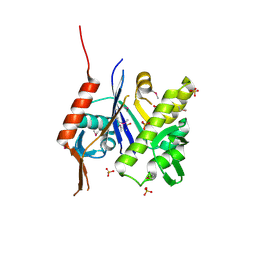 | | Crystal structure of B. subtilis TagU | | Descriptor: | GLYCEROL, Polyisoprenyl-teichoic acid--peptidoglycan teichoic acid transferase TagU, SULFATE ION | | Authors: | Li, F.K.K, Strynadka, N.C.J. | | Deposit date: | 2019-09-23 | | Release date: | 2020-01-29 | | Last modified: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic analysis ofStaphylococcus aureusLcpA, the primary wall teichoic acid ligase.
J.Biol.Chem., 295, 2020
|
|
3LCE
 
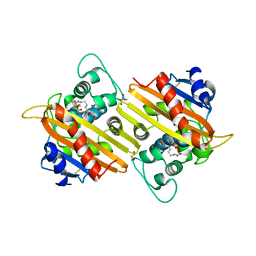 | | Crystal Structure of Oxa-10 Beta-Lactamase Covalently Bound to Cyclobutanone Beta-Lactam Mimic | | Descriptor: | (1S,3S,4S,5S)-7,7-dichloro-3-methoxy-2-thiabicyclo[3.2.0]heptan-6-one-4-carboxylic acid, Beta-lactamase OXA-10, GLYCEROL, ... | | Authors: | Gretes, M, Strynadka, N.C.J. | | Deposit date: | 2010-01-10 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cyclobutanone Analogues of beta-Lactams Revisited: Insights into Conformational Requirements for Inhibition of Serine- and Metallo-beta-Lactamases.
J.Am.Chem.Soc., 132, 2010
|
|
