5VFI
 
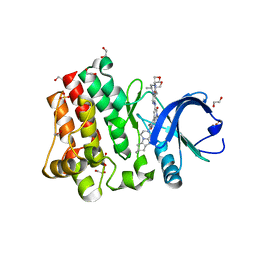 | | Bruton's tyrosine kinase (BTK) with GDC-0853 | | Descriptor: | 1,2-ETHANEDIOL, 2-[3'-(hydroxymethyl)-1-methyl-5-({5-[(2S)-2-methyl-4-(oxetan-3-yl)piperazin-1-yl]pyridin-2-yl}amino)-6-oxo[1,6-dihydro[3,4'-bipyridine]]-2'-yl]-7,7-dimethyl-3,4,7,8-tetrahydro-2H-cyclopenta[4,5]pyrrolo[1,2-a]pyrazin-1(6H)-one, SULFATE ION, ... | | Authors: | Steinbacher, S, Eigenbrot, C. | | Deposit date: | 2017-04-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Discovery of GDC-0853: A Potent, Selective, and Noncovalent Bruton's Tyrosine Kinase Inhibitor in Early Clinical Development.
J. Med. Chem., 61, 2018
|
|
1QSD
 
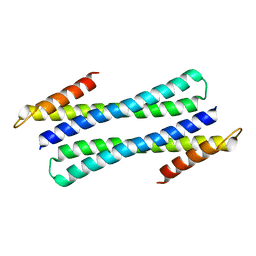 | | RBL2P, BETA-TUBULIN BINDING POST-CHAPERONIN COFACTOR | | Descriptor: | PROTEIN (BETA-TUBULIN BINDING POST-CHAPERONIN COFACTOR) | | Authors: | Steinbacher, S. | | Deposit date: | 1999-06-21 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the post-chaperonin beta-tubulin binding cofactor Rbl2p
Nat.Struct.Biol., 6, 1999
|
|
1U43
 
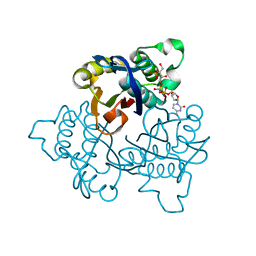 | | IspF with 4-diphosphocytidyl-2c-methyl-D-erythritol 2-phosphate | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 4-DIPHOSPHOCYTIDYL-2-C-METHYL-D-ERYTHRITOL 2-PHOSPHATE | | Authors: | Steinbacher, S, Kaiser, J, Wungsintaweekul, J, Hecht, S, Eisenreich, W, Gerhardt, S, Bacher, A, Rohdich, F. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of 2C-Methyl-D-Erythritol-2,4-Cyclodiphosphate Synthase Involved in Mevalonate Independent Biosynthesis of Isoprenoids
J.Mol.Biol., 316, 2002
|
|
1P0K
 
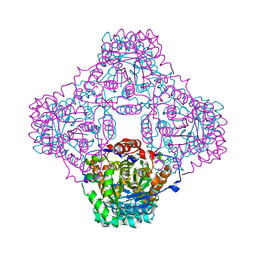 | | IPP:DMAPP isomerase type II apo structure | | Descriptor: | Isopentenyl-diphosphate delta-isomerase, PHOSPHATE ION | | Authors: | Steinbacher, S, Kaiser, J, Gerhardt, S, Eisenreich, W, Huber, R, Bacher, A, Rohdich, F. | | Deposit date: | 2003-04-10 | | Release date: | 2003-06-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Type II Isopentenyl Diphosphate:Dimethylallyl Diphosphate Isomerase from Bacillus subtilis
J.Mol.Biol., 329, 2003
|
|
1P0N
 
 | | IPP:DMAPP isomerase type II, FMN complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, Isopentenyl-diphosphate delta-isomerase | | Authors: | Steinbacher, S, Kaiser, J, Gerhardt, S, Eisenreich, W, Huber, R, Bacher, A, Rohdich, F. | | Deposit date: | 2003-04-10 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Type II Isopentenyl Diphosphate:Dimethylallyl Diphosphate Isomerase from Bacillus subtilis
J.Mol.Biol., 329, 2003
|
|
1U3P
 
 | | IspF native | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, ZINC ION | | Authors: | Steinbacher, S, Kaiser, J, Wungsintaweekul, J, Hecht, S, Eisenreich, W, Gerhardt, S, Bacher, A, Rohdich, F. | | Deposit date: | 2004-07-22 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of 2C-Methyl-D-Erythritol-2,4-Cyclodiphosphate Synthase Involved in Mevalonate Independent Biosynthesis of Isoprenoids
J.Mol.Biol., 316, 2002
|
|
1U40
 
 | | IspF with 4-diphosphocytidyl-2C-methyl-D-erythritol | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 4-DIPHOSPHOCYTIDYL-2-C-METHYL-D-ERYTHRITOL, ZINC ION | | Authors: | Steinbacher, S, Kaiser, J, Wungsintaweekul, J, Hecht, S, Eisenreich, W, Gerhardt, S, Bacher, A, Rohdich, F. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of 2C-Methyl-D-Erythritol-2,4-Cyclodiphosphate Synthase Involved in Mevalonate Independent Biosynthesis of Isoprenoids
J.Mol.Biol., 316, 2002
|
|
1U3L
 
 | | IspF with Mg and CDP | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Steinbacher, S, Kaiser, J, Wungsintaweekul, J, Hechst, S, Eisenreich, W, Gerhardt, S, Bacher, A, Rohdich, F. | | Deposit date: | 2004-07-22 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of 2C-Methyl-D-Erythritol-2,4-Cyclodiphosphate Synthase Involved in Mevalonate Independent Biosynthesis of Isoprenoids
J.Mol.Biol., 316, 2002
|
|
3JDW
 
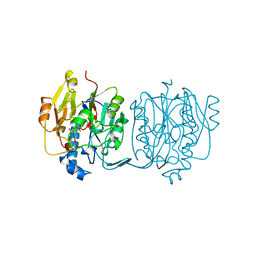 | | CRYSTAL STRUCTURE AND MECHANISM OF L-ARGININE: GLYCINE AMIDINOTRANSFERASE: A MITOCHONDRIAL ENZYME INVOLVED IN CREATINE BIOSYNTHESIS | | Descriptor: | L-ARGININE:GLYCINE AMIDINOTRANSFERASE, L-ornithine | | Authors: | Humm, A, Fritsche, E, Steinbacher, S, Huber, R. | | Deposit date: | 1997-01-24 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and mechanism of human L-arginine:glycine amidinotransferase: a mitochondrial enzyme involved in creatine biosynthesis.
EMBO J., 16, 1997
|
|
1QRB
 
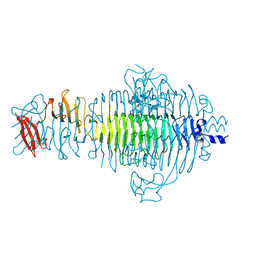 | | PLASTICITY AND STERIC STRAIN IN A PARALLEL BETA-HELIX: RATIONAL MUTATIONS IN P22 TAILSPIKE PROTEIN | | Descriptor: | PROTEIN (TAILSPIKE-PROTEIN) | | Authors: | Schuler, B, Furst, F, Osterroth, F, Steinbacher, S, Huber, R, Seckler, R. | | Deposit date: | 1999-06-12 | | Release date: | 2000-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Plasticity and steric strain in a parallel beta-helix: rational mutations in the P22 tailspike protein.
Proteins, 39, 2000
|
|
2JDW
 
 | |
4JDW
 
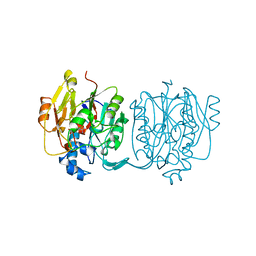 | | CRYSTAL STRUCTURE AND MECHANISM OF L-ARGININE: GLYCINE AMIDINOTRANSFERASE: A MITOCHONDRIAL ENZYME INVOLVED IN CREATINE BIOSYNTHESIS | | Descriptor: | ARGININE, L-ARGININE:GLYCINE AMIDINOTRANSFERASE | | Authors: | Humm, A, Fritsche, E, Steinbacher, S, Huber, R. | | Deposit date: | 1997-01-24 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and mechanism of human L-arginine:glycine amidinotransferase: a mitochondrial enzyme involved in creatine biosynthesis.
EMBO J., 16, 1997
|
|
3PLS
 
 | | RON in complex with ligand AMP-PNP | | Descriptor: | MAGNESIUM ION, Macrophage-stimulating protein receptor, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wang, J, Steinbacher, S, Augustin, M, Schreiner, P, Epstein, D, Mulvihill, M.J, Crew, A.P. | | Deposit date: | 2010-11-15 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Crystal Structure of a Constitutively Active Mutant RON Kinase Suggests an Intramolecular Autophosphorylation Hypothesis
Biochemistry, 49, 2010
|
|
4NMH
 
 | | 11-beta-HSD1 in complex with a 3,3-Di-methyl-azetidin-2-one | | Descriptor: | (4S)-4-(2-methoxyphenyl)-3,3-dimethyl-1-[3-(methylsulfonyl)phenyl]azetidin-2-one, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | McCoull, W, Augustin, M, Blake, C, Ertan, A, Kilgour, E.K, Krapp, S, Moore, J.E, Newcombe, N.J, Packer, M.J, Rees, A, Revill, J, Scott, J.S, Selmi, N, Gerhardt, S, Ogg, D.J, Steinbacher, S, Whittamore, P.R.O. | | Deposit date: | 2013-11-15 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification and optimisation of 3,3-dimethyl-azetidin-2-ones as potent and selective inhibitors of 11 beta-hydroxysteroid dehydrogenase type 1 (11-beta-HSD1)
TO BE PUBLISHED
|
|
2OAU
 
 | | Mechanosensitive Channel of Small Conductance (MscS) | | Descriptor: | Small-conductance mechanosensitive channel | | Authors: | Rees, D.C, Bass, R.B, Steinbacher, S, Strop, P, Barclay, M.T. | | Deposit date: | 2006-12-17 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structures of the Prokaryotic Mechanosensitive Channels MscL and MscS
CURRENT TOPICS IN MEMBRANES, 58, 2007
|
|
2OAR
 
 | | Mechanosensitive Channel of Large Conductance (MscL) | | Descriptor: | GOLD ION, Large-conductance mechanosensitive channel | | Authors: | Rees, D.C, Chang, G, Spencer, R.H, Lee, A.T, Steinbacher, S, Strop, P. | | Deposit date: | 2006-12-17 | | Release date: | 2007-01-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures of the Prokaryotic Mechanosensitive Channels MscL and MscS
Current Topics in Membranes, 58, 2007
|
|
7S25
 
 | | ROCK1 IN COMPLEX WITH LIGAND G4998 | | Descriptor: | 2-[3-(methoxymethyl)phenyl]-N-[4-(1H-pyrazol-4-yl)phenyl]acetamide, CHLORIDE ION, Rho-associated protein kinase 1 | | Authors: | Ganichkin, O, Harris, S.F, Steinbacher, S. | | Deposit date: | 2021-09-03 | | Release date: | 2022-10-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.337 Å) | | Cite: | Chemical space docking enables large-scale structure-based virtual screening to discover ROCK1 kinase inhibitors.
Nat Commun, 13, 2022
|
|
7S26
 
 | | ROCK1 IN COMPLEX WITH LIGAND G5018 | | Descriptor: | 2-[methyl(phenyl)amino]-1-[4-(1H-pyrrolo[2,3-b]pyridin-3-yl)-3,6-dihydropyridin-1(2H)-yl]ethan-1-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Rho-associated protein kinase 1 | | Authors: | Ganichkin, O, Harris, S.F, Steinbacher, S. | | Deposit date: | 2021-09-03 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.744 Å) | | Cite: | Chemical space docking enables large-scale structure-based virtual screening to discover ROCK1 kinase inhibitors.
Nat Commun, 13, 2022
|
|
3RO5
 
 | | Crystal structure of influenza A virus nucleoprotein with ligand | | Descriptor: | Nucleocapsid protein, [4-(2-chloro-4-nitrophenyl)piperazin-1-yl][3-(2-methoxyphenyl)-5-methyl-1,2-oxazol-4-yl]methanone | | Authors: | Pearce, B.C, Edavettal, S, McDonnell, P.A, Lewis, H.A, Steinbacher, S, Baldwin, E.T, Langley, D.R, Maskos, K, Mortl, M, Kiefersauer, R. | | Deposit date: | 2011-04-25 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Inhibition of influenza virus replication via small molecules that induce the formation of higher-order nucleoprotein oligomers.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6B8A
 
 | | Crystal structure of MvfR ligand binding domain in complex with M64 | | Descriptor: | 2-[(5-nitro-1H-benzimidazol-2-yl)sulfanyl]-N-(4-phenoxyphenyl)acetamide, COBALT HEXAMMINE(III), DNA-binding transcriptional regulator | | Authors: | Kitao, T, Steinbacher, S, Maskos, K, Blaesse, M, Rahme, L.G. | | Deposit date: | 2017-10-05 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Molecular Insights into Function and Competitive Inhibition ofPseudomonas aeruginosaMultiple Virulence Factor Regulator.
MBio, 9, 2018
|
|
8R7B
 
 | | SARS-CoV-2 NSP14 in complex with SAH and TDI-015051 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Guanine-N7 methyltransferase nsp14, ... | | Authors: | Meyer, C, Garzia, A, Miller, M, Huggins, D.J, Myers, R.W, Liverton, N, Kargman, S, Nitsche, J, Ganichkin, O, Steinbacher, S, Meinke, P.T, Tuschl, T. | | Deposit date: | 2023-11-24 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Small-molecule inhibition of SARS-CoV-2 guanine-N7 RNA cap methyltransferase
To Be Published
|
|
6AY2
 
 | | Structure of CathB with covalently linked Compound 28 | | Descriptor: | Cathepsin B, N~1~-[(2S)-1-amino-5-(carbamoylamino)pentan-2-yl]-N'~1~-[(1R)-1-(thiophen-3-yl)ethyl]cyclobutane-1,1-dicarboxamide | | Authors: | Kiefer, J.R, Steinbacher, S. | | Deposit date: | 2017-09-07 | | Release date: | 2017-12-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Peptidomimetic Antibody-Drug Conjugate Linkers with Enhanced Protease Specificity.
J. Med. Chem., 61, 2018
|
|
3TG6
 
 | | Crystal Structure of Influenza A Virus nucleoprotein with Ligand | | Descriptor: | Nucleocapsid protein, [4-(2-chloro-4-nitrophenyl)piperazin-1-yl][3-(2-chloropyridin-3-yl)-5-methyl-1,2-oxazol-4-yl]methanone | | Authors: | Pearce, B.C, Lewis, H.A, McDonnell, P.A, Steinbacher, S, Kiefersauer, R, Mortl, M, Maskos, K, Edavettal, S, Baldwin, E.T, Langley, D.R. | | Deposit date: | 2011-08-17 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Biophysical and Structural Characterization of a Novel Class of Influenza Virus Inhibitors
To be Published
|
|
7M2G
 
 | | INTERLEUKIN-2 (human) mutant P65K, C125S | | Descriptor: | GLYCEROL, Interleukin-2, SULFATE ION | | Authors: | Ptacin, J.L, Milla, M.E, Steinbacher, S, Maskos, K, Thomsen, M. | | Deposit date: | 2021-03-16 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | An engineered IL-2 reprogrammed for anti-tumor therapy using a semi-synthetic organism.
Nat Commun, 12, 2021
|
|
6H0E
 
 | | FAB dmCBTAU-22.1 IN COMPLEX WITH TAU PEPTIDE V1088-23 | | Descriptor: | GLYCEROL, HUMAN FAB ANTIBODY FRAGMENT OF dmCBTAU-22.1, Microtubule-associated protein tau | | Authors: | Juraszek, J, Steinbacher, S. | | Deposit date: | 2018-07-09 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Enhancement of therapeutic potential of a naturally occurring human antibody targeting a phosphorylated Ser422containing epitope on pathological tau.
Acta Neuropathol Commun, 6, 2018
|
|
