8EEB
 
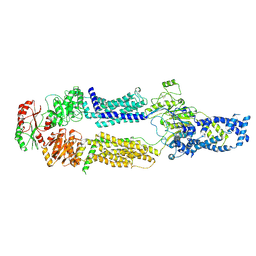 | | Cryo-EM structure of human ABCA7 in Digitonin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Phospholipid-transporting ATPase ABCA7, ... | | Authors: | Alam, A, Le, L.T.M, Thompson, J.R. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of human ABCA7 provide insights into its phospholipid translocation mechanisms.
Embo J., 42, 2023
|
|
8EOP
 
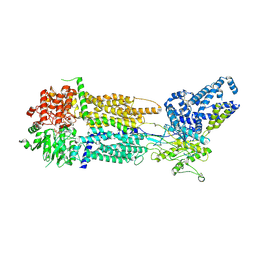 | | Cryo-EM Structure of Nanodisc reconstituted human ABCA7 EQ mutant in ATP bound closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Alam, A, Le, L.T.M, Thompson, J.R. | | Deposit date: | 2022-10-04 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of human ABCA7 provide insights into its phospholipid translocation mechanisms.
Embo J., 42, 2023
|
|
8EE6
 
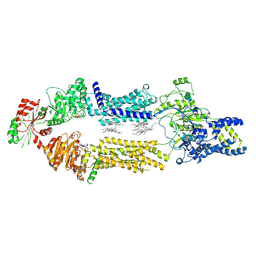 | | Cryo-EM Structure of human ABCA7 in PE/Ch nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Phospholipid-transporting ATPase ABCA7, ... | | Authors: | Alam, A, Le, L.T.M, Thompson, J.R. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of human ABCA7 provide insights into its phospholipid translocation mechanisms.
Embo J., 42, 2023
|
|
8EDW
 
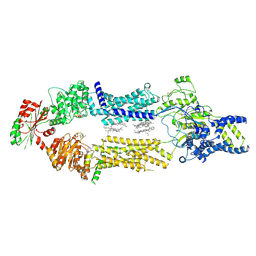 | | Cryo-EM Structure of human ABCA7 in BPL/Ch Nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Alam, A, Le, L.T.M, Thompson, J.R. | | Deposit date: | 2022-09-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of human ABCA7 provide insights into its phospholipid translocation mechanisms.
Embo J., 42, 2023
|
|
4ZIR
 
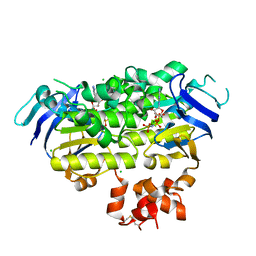 | | Crystal structure of EcfAA' heterodimer bound to AMPPNP | | Descriptor: | CHLORIDE ION, Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, ... | | Authors: | Karpowich, N.K, Cocco, N, Song, J.M, Wang, D.N. | | Deposit date: | 2015-04-28 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | ATP binding drives substrate capture in an ECF transporter by a release-and-catch mechanism.
Nat.Struct.Mol.Biol., 22, 2015
|
|
2LW8
 
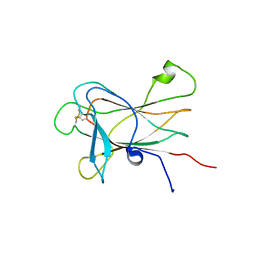 | |
8BIK
 
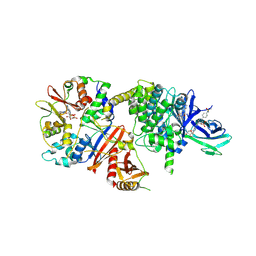 | | Crystal structure of human AMPK heterotrimer in complex with allosteric activator C455 | | Descriptor: | (3~{R},3~{a}~{R},6~{R},6~{a}~{R})-6-[[6-chloranyl-5-[4-[4-[[dimethyl(oxidanyl)-$l^{4}-sulfanyl]amino]phenyl]phenyl]-3~{H}-imidazo[4,5-b]pyridin-2-yl]oxy]-2,3,3~{a},5,6,6~{a}-hexahydrofuro[3,2-b]furan-3-ol, 5'-AMP-activated protein kinase catalytic subunit alpha-2, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Schimpl, M, Mather, K.M, Boland, M.L, Rivers, E.L, Srivastava, A, Hemsley, P, Robinson, J, Wan, P.T, Hansen, J, Read, J.A, Trevaskis, J.L, Smith, D.M. | | Deposit date: | 2022-11-02 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Direct beta 1/ beta 2 AMPK activation reduces liver steatosis but not fibrosis in a mouse model of non-alcoholic steatohepatitis
Biorxiv, 2024
|
|
7NAK
 
 | | Cryo-EM structure of activated human SARM1 in complex with NMN and 1AD (TIR:1AD) | | Descriptor: | NAD(+) hydrolase SARM1, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(5-iodanylisoquinolin-2-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Kerry, P.S, Nanson, J.D, Adams, S, Cunnea, K, Bosanac, T, Kobe, B, Hughes, R.O, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
7NAL
 
 | | Cryo-EM structure of activated human SARM1 in complex with NMN and 1AD (ARM and SAM domains) | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1 | | Authors: | Kerry, P.S, Nanson, J.D, Adams, S, Cunnea, K, Bosanac, T, Kobe, B, Hughes, R.O, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
7N0A
 
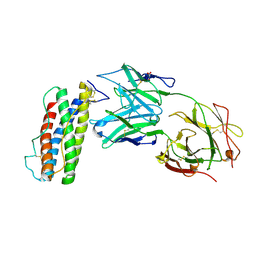 | | Structure of Human Leukaemia Inhibitory Factor with Fab MSC1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Leukemia inhibitory factor, ... | | Authors: | Raman, S, Bosch, A, Fransson, J, Julien, J.P. | | Deposit date: | 2021-05-25 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Therapeutic Targeting of LIF Overcomes Macrophage-mediated Immunosuppression of the Local Tumor Microenvironment.
Clin.Cancer Res., 29, 2023
|
|
7TVL
 
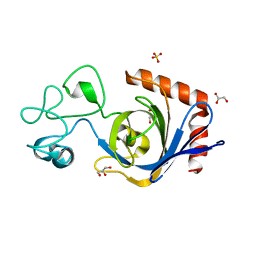 | | Viral AMG chitosanase V-Csn, apo structure | | Descriptor: | GLYCEROL, SULFATE ION, Viral chitosanase V-Csn | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
7TVN
 
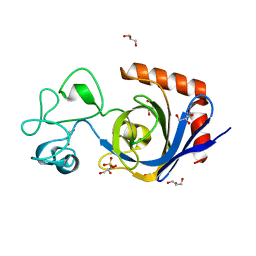 | | Viral AMG chitosanase V-Csn, D148N mutant | | Descriptor: | GLYCEROL, SULFATE ION, Viral chitosanase V-Csn D148N mutant | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
7TVO
 
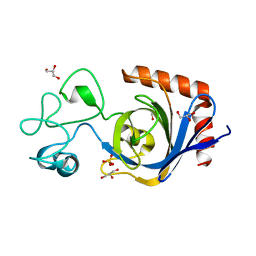 | | Viral AMG chitosanase V-Csn, E157Q mutant | | Descriptor: | GLYCEROL, SULFATE ION, Viral chitosanase V-Csn E157Q mutant | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
3IWE
 
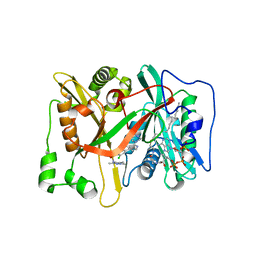 | | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD85646 | | Descriptor: | 2,6-dichloro-4-(2-piperazin-1-ylpyridin-4-yl)-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, Glycylpeptide N-tetradecanoyltransferase 1, TETRADECANOYL-COA | | Authors: | Qiu, W, Hutchinson, A, Wernimont, A, Lin, Y.-H, Kania, A, Ravichandran, M, Kozieradzki, I, Cossar, D, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Wyatt, P.G, Ferguson, M.A.J, Frearson, J.A, Brand, S.Y, Robinson, D.A, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-02 | | Release date: | 2009-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD85646
To be Published
|
|
6W8V
 
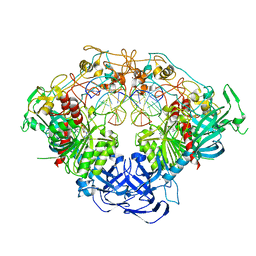 | | Crystal structure of mouse DNMT1 in complex with ACG DNA | | Descriptor: | ACG DNA (5'-D(*AP*CP*TP*TP*AP*(C49)P*GP*GP*AP*AP*GP*G)-3'), ACG DNA (5'-D(*CP*CP*TP*TP*CP*(5CM)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-21 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | DNMT1 activity, base flipping mechanism and genome-wide DNA methylation are regulated by the DNA sequence context
Nat Commun, 2020
|
|
6W8B
 
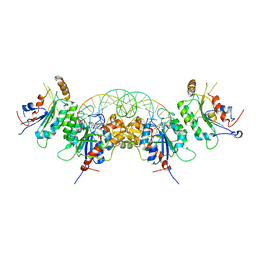 | | Structure of DNMT3A in complex with CGA DNA | | Descriptor: | CGA DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-20 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for impairment of DNA methylation by the DNMT3A R882H mutation.
Nat Commun, 11, 2020
|
|
3PCQ
 
 | | Femtosecond X-ray protein Nanocrystallography | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Chapman, H.N, Fromme, P, Barty, A, White, T.A, Kirian, R.A, Aquila, A, Hunter, M.S, Schulz, J, Deponte, D.P, Weierstall, U, Doak, R.B, Maia, F.R.N.C, Martin, A.V, Schlichting, I, Lomb, L, Coppola, N, Shoeman, R.L, Epp, S.W, Hartmann, R, Rolles, D, Rudenko, A, Foucar, L, Kimmel, N, Weidenspointner, G, Holl, P, Liang, M, Barthelmess, M, Caleman, C, Boutet, S, Bogan, M.J, Krzywinski, J, Bostedt, C, Bajt, S, Gumprecht, L, Rudek, B, Erk, B, Schmidt, C, Homke, A, Reich, C, Pietschner, D, Struder, L, Hauser, G, Gorke, H, Ullrich, J, Herrmann, S, Schaller, G, Schopper, F, Soltau, H, Kuhnel, K.-U, Messerschmidt, M, Bozek, J.D, Hau-Riege, S.P, Frank, M, Hampton, C.Y, Sierra, R, Starodub, D, Williams, G.J, Hajdu, J, Timneanu, N, Seibert, M.M, Andreasson, J, Rocker, A, Jonsson, O, Svenda, M, Stern, S, Nass, K, Andritschke, R, Schroter, C.-D, Krasniqi, F, Bott, M, Schmidt, K.E, Wang, X, Grotjohann, I, Holton, J.M, Barends, T.R.M, Neutze, R, Marchesini, S, Fromme, R, Schorb, S, Rupp, D, Adolph, M, Gorkhover, T, Andersson, I, Hirsemann, H, Potdevin, G, Graafsma, H, Nilsson, B, Spence, J.C.H. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (8.984 Å) | | Cite: | Femtosecond X-ray protein nanocrystallography.
Nature, 470, 2011
|
|
6W8W
 
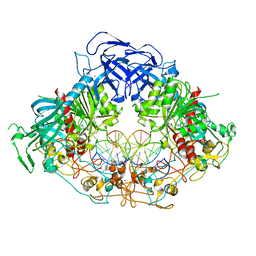 | | Crystal structure of mouse DNMT1 in complex with CCG DNA | | Descriptor: | CCG DNA (5'-D(*AP*CP*TP*TP*AP*(5CM)P*GP*GP*AP*AP*GP*G)-3'), CCG DNA (5'-D(*CP*CP*TP*TP*CP*(C49)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-21 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DNMT1 activity, base flipping mechanism and genome-wide DNA methylation are regulated by the DNA sequence context
Nat Commun, 2020
|
|
8OL8
 
 | |
6W89
 
 | | Structure of DNMT3A (R882H) in complex with CGA DNA | | Descriptor: | CGA DNA (25-MER), CITRIC ACID, DNA (cytosine-5)-methyltransferase 3-like, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-20 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structural basis for impairment of DNA methylation by the DNMT3A R882H mutation.
Nat Commun, 11, 2020
|
|
1O9S
 
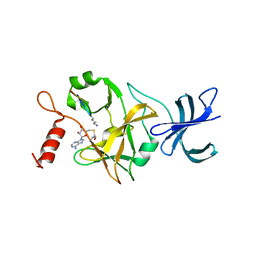 | | Crystal structure of a ternary complex of the human histone methyltransferase SET7/9 | | Descriptor: | GENE FRAGMENT FOR HISTONE H3, HISTONE-LYSINE N-METHYLTRANSFERASE, H3 LYSINE-4 SPECIFIC, ... | | Authors: | Xiao, B, Jing, C, Wilson, J.R, Walker, P.A, Vasisht, N, Kelly, G, Howell, S, Taylor, I.A, Blackburn, G.M, Gamblin, S.J. | | Deposit date: | 2002-12-18 | | Release date: | 2003-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Catalytic Mechanism of the Human Histone Methyltransferase Set7/9
Nature, 421, 2003
|
|
3IU1
 
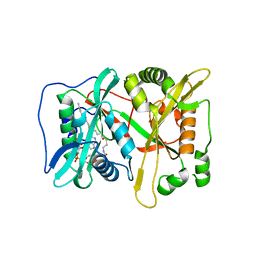 | | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA | | Descriptor: | Glycylpeptide N-tetradecanoyltransferase 1, TETRADECANOYL-COA | | Authors: | Qiu, W, Hutchinson, A, Wernimont, A, Lin, Y.-H, Kania, A, Ravichandran, M, Kozieradzki, I, Cossar, D, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Wyatt, P.G, Ferguson, M.A.J, Frearson, J.A, Brand, S.Y, Robinson, D.A, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-29 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA
To be Published
|
|
6W8J
 
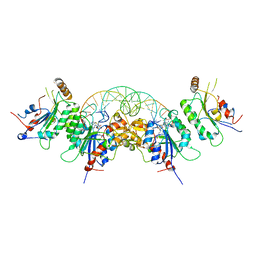 | | Structure of DNMT3A (R882H) in complex with CAG DNA | | Descriptor: | CAG DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-20 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Structural basis for impairment of DNA methylation by the DNMT3A R882H mutation.
Nat Commun, 11, 2020
|
|
6W8D
 
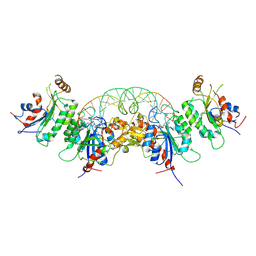 | | Structure of DNMT3A (R882H) in complex with CGT DNA | | Descriptor: | CGT DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, ... | | Authors: | Anteneh, H, Song, J. | | Deposit date: | 2020-03-20 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Structural basis for impairment of DNA methylation by the DNMT3A R882H mutation.
Nat Commun, 11, 2020
|
|
6O3A
 
 | | Crystal structure of Frizzled 7 CRD in complex with F7.B Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ... | | Authors: | Raman, S, Beilschmidt, M, Fransson, J, Julien, J.P. | | Deposit date: | 2019-02-26 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-guided design fine-tunes pharmacokinetics, tolerability, and antitumor profile of multispecific frizzled antibodies.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
