6QYL
 
 | | Structure of MBP-Mcl-1 in complex with compound 8a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QYK
 
 | | Structure of MBP-Mcl-1 in complex with compound 7a | | Descriptor: | (2~{R})-2-[6-ethyl-5-(1~{H}-indol-5-yl)thieno[2,3-d]pyrimidin-4-yl]oxypropanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QZ6
 
 | | Structure of Mcl-1 in complex with compound 8b | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QYN
 
 | | Structure of MBP-Mcl-1 in complex with compound 10d | | Descriptor: | (2~{R})-2-[5-(3-chloranyl-2-methyl-4-oxidanyl-phenyl)-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QYP
 
 | | Structure of Mcl-1 in complex with compound 13 | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-5-(4-methylpiperazin-1-yl)-4-oxidanyl-phenyl]-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QZ5
 
 | | Structure of Mcl-1 in complex with compound 8a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QYO
 
 | | Structure of MBP-Mcl-1 in complex with compound 18a | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QZ7
 
 | | Structure of MBP-Mcl-1 in complex with compound 8b | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QZB
 
 | | Structure of Mcl-1 in complex with compound 8d | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(2-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QZ8
 
 | | Structure of Mcl-1 in complex with compound 10d | | Descriptor: | (2~{R})-2-[5-(3-chloranyl-2-methyl-4-oxidanyl-phenyl)-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
6QXJ
 
 | | Structure of MBP-Mcl-1 in complex with compound 6a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-5-yl)thieno[2,3-d]pyrimidin-4-yl]amino]propanoic acid, Maltose-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-07 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
3EOC
 
 | | Cdk2/CyclinA complexed with a imidazo triazin-2-amine | | Descriptor: | 5-methyl-7-phenyl-N-(3,4,5-trimethoxyphenyl)imidazo[5,1-f][1,2,4]triazin-2-amine, Cell division protein kinase 2, Cyclin-A2 | | Authors: | Cheung, M, Kuntz, K, Pobanz, M, Salovich, J, Wilson, B, Andrews, W, Shewchuk, L, Epperly, A, Hassler, D, Leesnitzer, M, Smith, J, Smith, G, Lansing, T, Mook, R. | | Deposit date: | 2008-09-26 | | Release date: | 2008-11-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Imidazo[5,1-f][1,2,4]triazin-2-amines as novel inhibitors of polo-like kinase 1.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2H7X
 
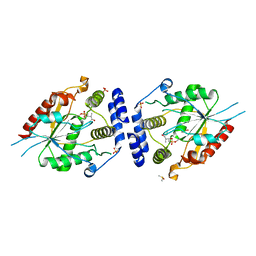 | | Pikromycin Thioesterase adduct with reduced triketide affinity label | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Giraldes, J.W, Akey, D.L, Kittendorf, J.D, Sherman, D.H, Smith, J.S, Fecik, R.A. | | Deposit date: | 2006-06-05 | | Release date: | 2006-09-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Mechanistic Insights of Polyketide Macrolactonization from Polyketide-based Affinity Labels
NAT.CHEM.BIOL., 2, 2006
|
|
2H7Y
 
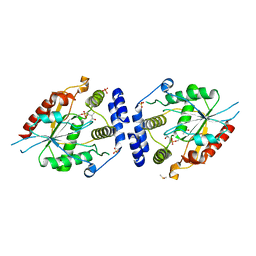 | | Pikromycin Thioesterase with covalent affinity label | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Giraldes, J.W, Akey, D.L, Kittendorf, J.D, Sherman, D.H, Smith, J.S, Fecik, R.A. | | Deposit date: | 2006-06-06 | | Release date: | 2006-09-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mechanistic Insights of Polyketide Macrolactonization from Polyketide-based Affinity Labels
NAT.CHEM.BIOL., 2, 2006
|
|
3NWT
 
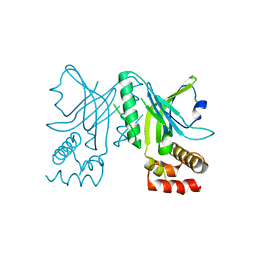 | | Crystal structure of the N-terminal domain of the yeast telomere-binding and telomerase regulatory protein Cdc13 | | Descriptor: | Cell division control protein 13 | | Authors: | Mitchell, M.T, Smith, J.S, Mason, M, Harper, S, Speicher, D.W, Johnson, F.B, Skordalakes, E. | | Deposit date: | 2010-07-10 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cdc13 N-terminal dimerization, DNA binding, and telomere length regulation.
Mol.Cell.Biol., 30, 2010
|
|
1FBZ
 
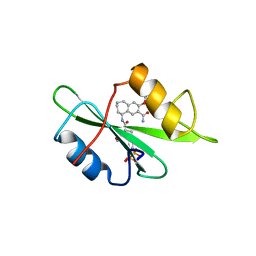 | | Structure-based design of a novel, osteoclast-selective, nonpeptide Src SH2 inhibitor with in vivo anti-resorptive activity | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE LCK, {4-[2-ACETYLAMINO-2-(3-CARBAMOYL-2-CYCLOHEXYLMETHOXY-6,7,8,9-TETRAHYDRO-5H-BENZOCYCLOHEPTEN-5YLCARBAMOYL)-ETHYL]-2-PHOSPHONO-PHENYL}-PHOSPHONIC ACID | | Authors: | Shakespeare, W, Yang, M, Bohacek, R, Cerasoli, F, Stebbis, K, Sundaramoorthi, R, Vu, C, Pradeepan, S, Metcalf, C, Haraldson, C, Merry, T, Dalgarno, D, Narula, S, Hatada, M, Lu, X, Van Schravendijk, M.R, Adams, S, Violette, S, Smith, J, Guan, W, Bartlett, C, Herson, J, Iuliucci, J, Weigele, M, Sawyer, T. | | Deposit date: | 2000-07-17 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design of an osteoclast-selective, nonpeptide src homology 2 inhibitor with in vivo antiresorptive activity.
Proc.Natl.Acad.Sci.Usa, 97, 2000
|
|
3NWS
 
 | | Crystal structure of the N-terminal domain of the yeast telomere-binding and telomerase regulatory protein Cdc13 | | Descriptor: | Cell division control protein 13 | | Authors: | Mitchell, M.T, Smith, J.S, Mason, M, Harper, S, Speicher, D.W, Johnson, F.B, Skordalakes, E. | | Deposit date: | 2010-07-10 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cdc13 N-terminal dimerization, DNA binding, and telomere length regulation.
Mol.Cell.Biol., 30, 2010
|
|
2HLI
 
 | | Solution structure of Crotonaldehyde-Derived N2-[3-Oxo-1(S)-methyl-propyl]-dG DNA Adduct in the 5'-CpG-3' Sequence | | Descriptor: | DNA dodecamer, DNA dodecamer with S-crotonaldehyde adduct | | Authors: | Cho, Y.-J, Wang, H, Kozekov, I.D, Kurtz, A.J, Jacob, J, Voehler, M, Smith, J, Harris, T.M, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2006-07-07 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Orientation of the Crotonaldehyde-Derived N(2)-[3-Oxo-1(S)-methyl-propyl]-dG DNA Adduct Hinders Interstrand Cross-Link Formation in the 5'-CpG-3' Sequence
Chem.Res.Toxicol., 19, 2006
|
|
1CNP
 
 | | THE STRUCTURE OF CALCYCLIN REVEALS A NOVEL HOMODIMERIC FOLD FOR S100 CA2+-BINDING PROTEINS, NMR, 22 STRUCTURES | | Descriptor: | CALCYCLIN (RABBIT, APO) | | Authors: | Potts, B.C.M, Smith, J, Akke, M, Macke, T.J, Okazaki, K, Hidaka, H, Case, D.A, Chazin, W.J. | | Deposit date: | 1995-08-31 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of calcyclin reveals a novel homodimeric fold for S100 Ca(2+)-binding proteins.
Nat.Struct.Biol., 2, 1995
|
|
2NDJ
 
 | | Structural Basis for KCNE3 and Estrogen Modulation of the KCNQ1 Channel | | Descriptor: | Potassium voltage-gated channel subfamily E member 3 | | Authors: | Sanders, C.R, Van Horn, W.D, Kroncke, B.M, Sisco, N.J, Meiler, J, Vanoye, C.G, Song, Y, Nannemann, D.P, Welch, R.C, Kang, C, Smith, J, George, A.L. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for KCNE3 modulation of potassium recycling in epithelia.
Sci Adv, 2, 2016
|
|
7MRX
 
 | |
2XVO
 
 | | SSO1725, a protein involved in the CRISPR/Cas pathway | | Descriptor: | BETA-MERCAPTOETHANOL, SSO1725, SULFATE ION | | Authors: | Reeks, J, Liu, H, Naismith, J, White, M, McMahon, S. | | Deposit date: | 2010-10-26 | | Release date: | 2010-12-29 | | Last modified: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure and Mechanism of the Cmr Complex for Crispr-Mediated Antiviral Immunity.
Mol.Cell, 45, 2012
|
|
5FNV
 
 | | a new complex structure of tubulin with an alpha-beta unsaturated lactone | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, Y, Naismith, J, Zhu, X. | | Deposit date: | 2015-11-16 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Pironetin Reacts Covalently with Cysteine-316 of Alpha-Tubulin to Destabilize Microtubule.
Nat.Commun., 7, 2016
|
|
5O78
 
 | |
5O9C
 
 | |
