6BX3
 
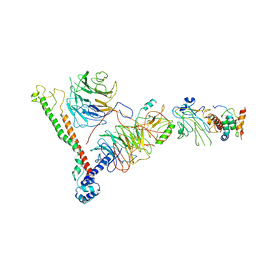 | | Structure of histone H3k4 methyltransferase | | Descriptor: | COMPASS component BRE2, COMPASS component SDC1, COMPASS component SPP1, ... | | Authors: | Skiniotis, G, Qu, Q.H. | | Deposit date: | 2017-12-16 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and Conformational Dynamics of a COMPASS Histone H3K4 Methyltransferase Complex.
Cell, 174, 2018
|
|
6WHA
 
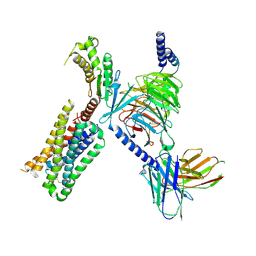 | | HTR2A bound to 25-CN-NBOH in complex with a mini-Galpha-q protein, beta/gamma subunits and an active-state stabilizing single-chain variable fragment (scFv16) obtained by cryo-electron microscopy (cryoEM) | | Descriptor: | 4-(2-{[(2-hydroxyphenyl)methyl]amino}ethyl)-2,5-dimethoxybenzonitrile, G subunit q (Gi2-mini-Gq chimeric), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Skiniotis, G, Roth, B, Kim, K, Panova, O. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structure of a Hallucinogen-Activated Gq-Coupled 5-HT2A Serotonin Receptor
Cell(Cambridge,Mass.), 182, 2020
|
|
6DDE
 
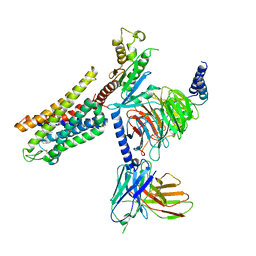 | | Mu Opioid Receptor-Gi Protein Complex | | Descriptor: | DAMGO, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Koehl, A, Hu, H, Maeda, S, Manglik, A, Zhang, Y, Kobilka, B.K, Skiniotis, G, Weis, W.I. | | Deposit date: | 2018-05-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the mu-opioid receptor-Giprotein complex.
Nature, 558, 2018
|
|
6U1N
 
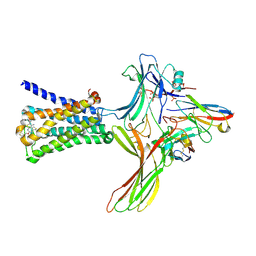 | | GPCR-Beta arrestin structure in lipid bilayer | | Descriptor: | 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno[2,3-b]pyridine-2-carboxamide, Beta-arrestin-1, Fab30 heavy chain, ... | | Authors: | Staus, D.P, Hu, H, Robertson, M.J, Kleinhenz, A.L.W, Wingler, L.M, Capel, W.D, Latorraca, N.R, Lefkowitz, R.J, Skiniotis, G. | | Deposit date: | 2019-08-16 | | Release date: | 2020-02-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the M2 muscarinic receptor-beta-arrestin complex in a lipid nanodisc.
Nature, 579, 2020
|
|
8V6U
 
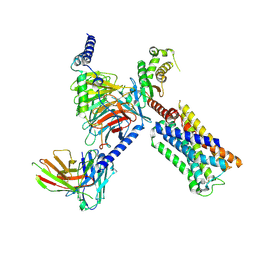 | | 5HT2AR-miniGq heterotrimer in complex with a novel agonist obtained from large scale docking | | Descriptor: | 4-{(3R)-1-[(1R)-1-(pyrimidin-2-yl)ethyl]piperidin-3-yl}phenol, 5-hydroxytryptamine receptor 2A, G protein alpha-subunit q (Gi2-mini-Gq chimera), ... | | Authors: | Gumpper, R.H, Wang, L, Kapolka, N, Skiniotis, G, Roth, B.L. | | Deposit date: | 2023-12-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | AlphaFold2 structures guide prospective ligand discovery.
Science, 384, 2024
|
|
9C1P
 
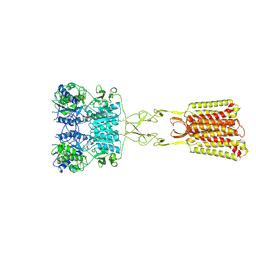 | | Structure of Calcium-Sensing Receptor in complex with positive allosteric modulator '6218 | | Descriptor: | (5R)-N-[2-(1,2-benzothiazol-3-yl)ethyl]-1-methyl-2,3,4,5-tetrahydro-1H-1-benzazepin-5-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wu, C, Skiniotis, G. | | Deposit date: | 2024-05-29 | | Release date: | 2024-10-02 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Large library docking identifies positive allosteric modulators of the calcium-sensing receptor.
Science, 385, 2024
|
|
9C2F
 
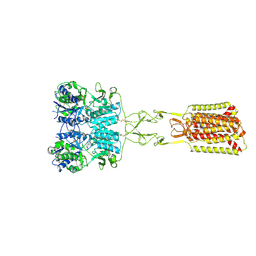 | | Structure of Calcium-Sensing Receptor in complex with positive allosteric modulator '54149 | | Descriptor: | (1R)-1-(2H-1,3-benzodioxol-4-yl)-N-[2-(1,2-benzothiazol-3-yl)ethyl]ethan-1-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, C, Skiniotis, G. | | Deposit date: | 2024-05-30 | | Release date: | 2024-10-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Large library docking identifies positive allosteric modulators of the calcium-sensing receptor.
Science, 385, 2024
|
|
3JCS
 
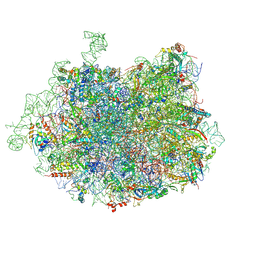 | | 2.8 Angstrom cryo-EM structure of the large ribosomal subunit from the eukaryotic parasite Leishmania | | Descriptor: | 26S alpha ribosomal RNA, 26S delta ribosomal RNA, 26S epsilon ribosomal RNA, ... | | Authors: | Shalev-Benami, M, Zhang, Y, Matzov, D, Halfon, Y, Zackay, A, Rozenberg, H, Zimmerman, E, Bashan, A, Jaffe, C.L, Yonath, A, Skiniotis, G. | | Deposit date: | 2016-01-21 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | 2.8- angstrom Cryo-EM Structure of the Large Ribosomal Subunit from the Eukaryotic Parasite Leishmania.
Cell Rep, 16, 2016
|
|
3SN6
 
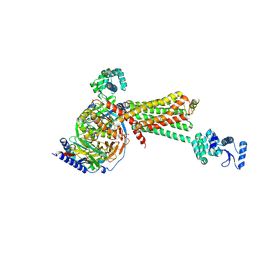 | | Crystal structure of the beta2 adrenergic receptor-Gs protein complex | | Descriptor: | 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Camelid antibody VHH fragment, Endolysin,Beta-2 adrenergic receptor, ... | | Authors: | Rasmussen, S.G.F, DeVree, B.T, Zou, Y, Kruse, A.C, Chung, K.Y, Kobilka, T.S, Thian, F.S, Chae, P.S, Pardon, E, Calinski, D, Mathiesen, J.M, Shah, S.T.A, Lyons, J.A, Caffrey, M, Gellman, S.H, Steyaert, J, Skiniotis, G, Weis, W.I, Sunahara, R.K, Kobilka, B.K. | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the beta2 adrenergic receptor-Gs protein complex
Nature, 477, 2011
|
|
8EL8
 
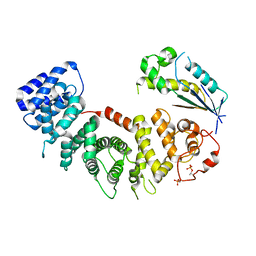 | |
8EL7
 
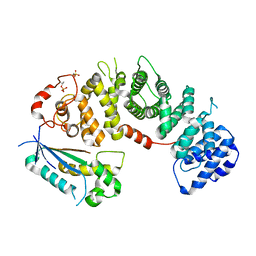 | |
6OIJ
 
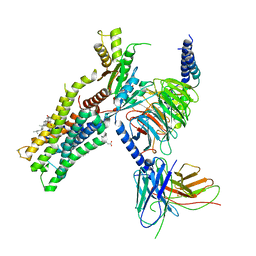 | | Muscarinic acetylcholine receptor 1-G11 protein complex | | Descriptor: | 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, Antibody fragment, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Maeda, S, Qianhui, Q, Skiniotis, G, Kobilka, B. | | Deposit date: | 2019-04-09 | | Release date: | 2019-05-08 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of the M1 and M2 muscarinic acetylcholine receptor/G-protein complexes.
Science, 364, 2019
|
|
6OYA
 
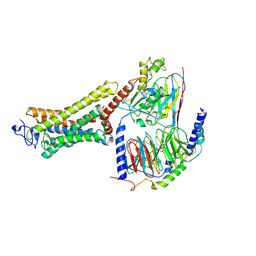 | | Structure of the Rhodopsin-Transducin-Nanobody Complex | | Descriptor: | Camelid antibody VHH fragment, Gt-alpha/Gi1-alpha chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Gao, Y, Hu, H, Ramachandran, S, Erickson, J.W, Cerione, R.A, Skiniotis, G. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of the Rhodopsin-Transducin Complex: Insights into G-Protein Activation.
Mol.Cell, 75, 2019
|
|
6OSA
 
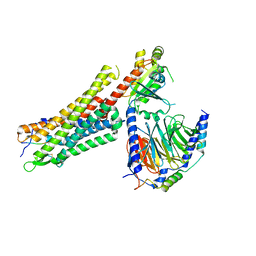 | | human Neurotensin Receptor 1 (hNTSR1) - Gi1 Protein Complex in non-canonical conformation (NC state) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Kato, H.E, Zhang, Y, Kobilka, B.K, Skiniotis, G. | | Deposit date: | 2019-05-01 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational transitions of a neurotensin receptor 1-Gi1complex.
Nature, 572, 2019
|
|
7M3G
 
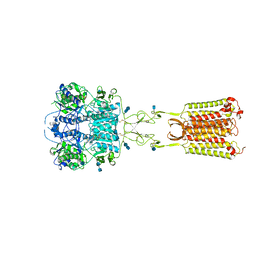 | | Asymmetric Activation of the Calcium Sensing Receptor Homodimer | | Descriptor: | 2-[4-[(3S)-3-[[(1R)-1-naphthalen-1-ylethyl]amino]pyrrolidin-1-yl]phenyl]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, Y, Robertson, M.J, Zhang, C, Meyerowitz, J.G, Panova, O, Skiniotis, G. | | Deposit date: | 2021-03-18 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Asymmetric activation of the calcium-sensing receptor homodimer.
Nature, 595, 2021
|
|
7M3J
 
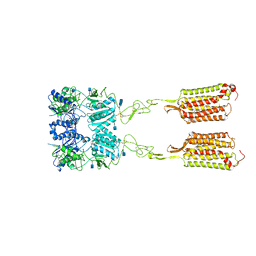 | | Asymmetric Activation of the Calcium Sensing Receptor Homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-chloro-6-[(2R)-2-hydroxy-3-{[2-methyl-1-(naphthalen-2-yl)propan-2-yl]amino}propoxy]benzonitrile, ... | | Authors: | Gao, Y, Robertson, M.J, Zhang, C, Meyerowitz, J.G, Panova, O, Skiniotis, G. | | Deposit date: | 2021-03-18 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Asymmetric activation of the calcium-sensing receptor homodimer.
Nature, 595, 2021
|
|
7M3E
 
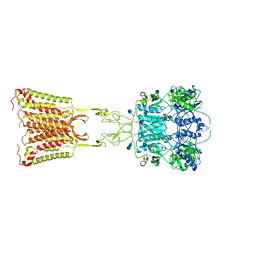 | | Asymmetric Activation of the Calcium Sensing Receptor Homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-chloro-6-[(2R)-2-hydroxy-3-{[2-methyl-1-(naphthalen-2-yl)propan-2-yl]amino}propoxy]benzonitrile, ... | | Authors: | Gao, Y, Robertson, M.J, Zhang, C, Meyerowitz, J.G, Panova, O, Skiniotis, G. | | Deposit date: | 2021-03-18 | | Release date: | 2021-06-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Asymmetric activation of the calcium-sensing receptor homodimer.
Nature, 595, 2021
|
|
7JSN
 
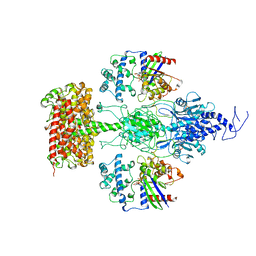 | | Structure of the Visual Signaling Complex between Transducin and Phosphodiesterase 6 | | Descriptor: | 2-{2-ETHOXY-5-[(4-ETHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-5-METHYL-7-PROPYLIMIDAZO[5,1-F][1,2,4]TRIAZIN-4(1H)-ONE, GUANOSINE-3',5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gao, Y, Eskici, G, Ramachandran, S, Skiniotis, G, Cerione, R.A. | | Deposit date: | 2020-08-15 | | Release date: | 2020-10-21 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the Visual Signaling Complex between Transducin and Phosphodiesterase 6.
Mol.Cell, 80, 2020
|
|
4X8P
 
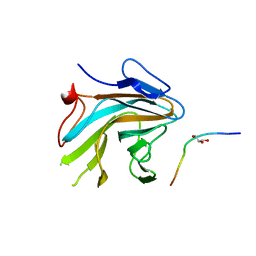 | | Crystal structure of Ash2L SPRY domain in complex with RbBP5 | | Descriptor: | GLYCEROL, Retinoblastoma-binding protein 5, Set1/Ash2 histone methyltransferase complex subunit ASH2,Set1/Ash2 histone methyltransferase complex subunit ASH2 | | Authors: | Zhang, P, Chaturvedi, C.P, Brunzelle, J.S, Skiniotis, G, Brand, M, Shilatifard, A, Couture, J.-F. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A phosphorylation switch on RbBP5 regulates histone H3 Lys4 methylation.
Genes Dev., 29, 2015
|
|
4X8N
 
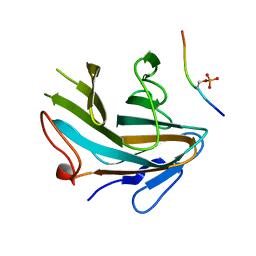 | | Crystal structure of Ash2L SPRY domain in complex with phosphorylated RbBP5 | | Descriptor: | Retinoblastoma-binding protein 5, Set1/Ash2 histone methyltransferase complex subunit ASH2 | | Authors: | Zhang, P, Chaturvedi, C.P, Brunzelle, J.S, Skiniotis, G, Brand, M, Shilatifard, A, Couture, J.-F. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A phosphorylation switch on RbBP5 regulates histone H3 Lys4 methylation.
Genes Dev., 29, 2015
|
|
6DDF
 
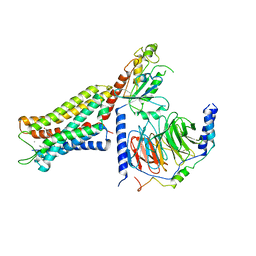 | | Mu Opioid Receptor-Gi Protein Complex | | Descriptor: | DAMGO, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Koehl, A, Hu, H, Maeda, S, Manglik, A, Kobilka, B.K, Skiniotis, G, Weis, W.I. | | Deposit date: | 2018-05-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the mu-opioid receptor-Giprotein complex.
Nature, 558, 2018
|
|
7JHJ
 
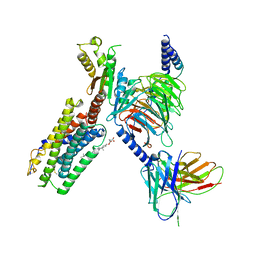 | | Structure of the Epstein-Barr virus GPCR BILF1 in complex with human Gi | | Descriptor: | Antibody fragment scFv16, BILF1, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Tsutsumi, N, Qu, Q.H, Skiniotis, G, Garcia, K.C. | | Deposit date: | 2020-07-20 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for the constitutive activity and immunomodulatory properties of the Epstein-Barr virus-encoded G protein-coupled receptor BILF1.
Immunity, 54, 2021
|
|
6OS9
 
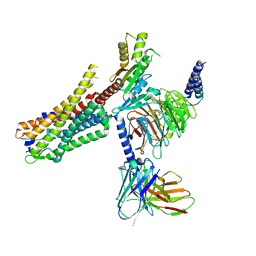 | | human Neurotensin Receptor 1 (hNTSR1) - Gi1 Protein Complex in canonical conformation (C state) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Kato, H.E, Zhang, Y, Kobilka, B.K, Skiniotis, G. | | Deposit date: | 2019-05-01 | | Release date: | 2019-07-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Conformational transitions of a neurotensin receptor 1-Gi1complex.
Nature, 572, 2019
|
|
6OY9
 
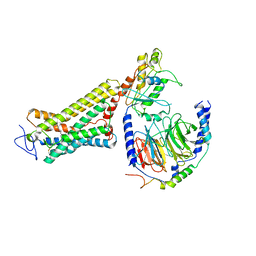 | | Structure of the Rhodopsin-Transducin Complex | | Descriptor: | Gt-alpha/Gi1-alpha chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, ... | | Authors: | Gao, Y, Hu, H, Ramachandran, S, Erickson, J.W, Cerione, R.A, Skiniotis, G. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of the Rhodopsin-Transducin Complex: Insights into G-Protein Activation.
Mol.Cell, 75, 2019
|
|
6OIK
 
 | | Muscarinic acetylcholine receptor 2-Go complex | | Descriptor: | 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno[2,3-b]pyridine-2-carboxamide, 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, Antibody fragment, ... | | Authors: | Maeda, S, Qianhui, Q, Skiniotis, G, Kobilka, B. | | Deposit date: | 2019-04-09 | | Release date: | 2019-05-08 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of the M1 and M2 muscarinic acetylcholine receptor/G-protein complexes.
Science, 364, 2019
|
|
