7XLY
 
 | | Crystal structure of FadA2 (Rv0243) from the fatty acid metabolic pathway of Mycobacterium tuberculosis | | Descriptor: | Probable acetyl-CoA acyltransferase FadA2 (3-ketoacyl-CoA thiolase) (Beta-ketothiolase), SULFATE ION | | Authors: | Singh, R, Kundu, P, Singh, B.K, Bhattacharyya, S, Das, A.K. | | Deposit date: | 2022-04-23 | | Release date: | 2023-04-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of FadA2 thiolase from Mycobacterium tuberculosis and prediction of its substrate specificity and membrane-anchoring properties.
Febs J., 290, 2023
|
|
7BKP
 
 | | CryoEM structure of disease related M854K MDA5-dsRNA filament in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, MAGNESIUM ION, ... | | Authors: | Singh, R, Herrero del Valle, A, Yu, Q, Modis, Y. | | Deposit date: | 2021-01-16 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | MDA5 disease variant M854K prevents ATP-dependent structural discrimination of viral and cellular RNA.
Nat Commun, 12, 2021
|
|
6GYF
 
 | | Crystal structure of NadR protein in complex with NR | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, MAGNESIUM ION, Nicotinamide-nucleotide adenylyltransferase NadR family / Ribosylnicotinamide kinase, ... | | Authors: | Singh, R, Stetsenko, A, Jaehme, M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2018-06-29 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Characterization of NadR fromLactococcus lactis.
Molecules, 25, 2020
|
|
6GYE
 
 | | Crystal structure of NadR protein in complex with NR | | Descriptor: | Nicotinamide riboside, Nicotinamide-nucleotide adenylyltransferase NadR family / Ribosylnicotinamide kinase, SULFATE ION | | Authors: | Singh, R, Stetsenko, A, Jaehme, M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2018-06-29 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Characterization of NadR fromLactococcus lactis.
Molecules, 25, 2020
|
|
6GZO
 
 | | Crystal structure of NadR protein in complex with NAD and AMP-PNP | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Nicotinamide-nucleotide adenylyltransferase NadR family / Ribosylnicotinamide kinase, PHOSPHATE ION, ... | | Authors: | Singh, R, Stetsenko, A, Jaehme, M, Guskov, A, Slotboom, D.J. | | Deposit date: | 2018-07-04 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Characterization of NadR fromLactococcus lactis.
Molecules, 25, 2020
|
|
7YVY
 
 | |
2F2K
 
 | | Aldose reductase tertiary complex with NADPH and DEG | | Descriptor: | Aldose reductase, GAMMA-GLUTAMYL-S-(1,2-DICARBOXYETHYL)CYSTEINYLGLYCINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Singh, R, White, M.A, Ramana, K.V, Petrash, J.M, Watowich, S.J, Bhatnagar, A, Srivastava, S.K. | | Deposit date: | 2005-11-17 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure of a glutathione conjugate bound to the active site of aldose reductase.
Proteins, 64, 2006
|
|
3QNR
 
 | | DyPB from Rhodococcus jostii RHA1, crystal form 1 | | Descriptor: | DyP Peroxidase, FORMIC ACID, GLYCEROL, ... | | Authors: | Singh, R, Roberts, J.N, Grigg, J.C, Eltis, L.D, Murphy, M.E.P. | | Deposit date: | 2011-02-09 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characterization of dye-decolorizing peroxidases from Rhodococcus jostii RHA1.
Biochemistry, 50, 2011
|
|
6A9U
 
 | | Crystal strcture of Icp55 from Saccharomyces cerevisiae bound to apstatin inhibitor | | Descriptor: | Intermediate cleaving peptidase 55, MANGANESE (II) ION, apstatin | | Authors: | Singh, R, Kumar, A, Goyal, V.D, Makde, R.D. | | Deposit date: | 2018-07-16 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures and biochemical analyses of intermediate cleavage peptidase: role of dynamics in enzymatic function.
FEBS Lett., 593, 2019
|
|
6A9V
 
 | | Crystal structure of Icp55 from Saccharomyces cerevisiae (N-terminal 42 residues deletion) | | Descriptor: | GLYCINE, Intermediate cleaving peptidase 55, MANGANESE (II) ION, ... | | Authors: | Singh, R, Kumar, A, Goyal, V.D, Makde, R.D. | | Deposit date: | 2018-07-16 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures and biochemical analyses of intermediate cleavage peptidase: role of dynamics in enzymatic function.
FEBS Lett., 593, 2019
|
|
5X49
 
 | | Crystal Structure of Human mitochondrial X-prolyl Aminopeptidase (XPNPEP3) | | Descriptor: | (2S,3R)-3-amino-2-hydroxy-4-phenylbutanoic acid, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Singh, R, Kumar, A, Ghosh, B, Jamdar, S, Makde, R.D. | | Deposit date: | 2017-02-10 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the human aminopeptidase XPNPEP3 and comparison of its in vitro activity with Icp55 orthologs: Insights into diverse cellular processes.
J. Biol. Chem., 292, 2017
|
|
6A9T
 
 | | Crystal structure of Icp55 from Saccharomyces cerevisiae (N-terminal 58 residues deletion) | | Descriptor: | GLYCINE, Intermediate cleaving peptidase 55, MANGANESE (II) ION, ... | | Authors: | Singh, R, Kumar, A, Goyal, V.D, Makde, R.D. | | Deposit date: | 2018-07-16 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures and biochemical analyses of intermediate cleavage peptidase: role of dynamics in enzymatic function.
FEBS Lett., 593, 2019
|
|
7CAY
 
 | | Crystal Structure of Lon N-terminal domain protein from Xanthomonas campestris | | Descriptor: | ATP-dependent protease | | Authors: | Singh, R, Sharma, B, Deshmukh, S, Kumar, A, Makde, R.D. | | Deposit date: | 2020-06-10 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of XCC3289 from Xanthomonas campestris: homology with the N-terminal substrate-binding domain of Lon peptidase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
3ZJT
 
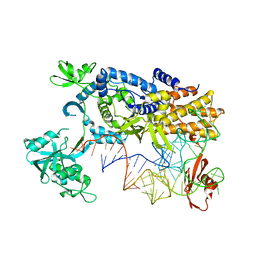 | | Ternary complex of E.coli leucyl-tRNA synthetase, tRNA(Leu)574 and the benzoxaborole AN3017 in the editing conformation | | Descriptor: | LEUCYL-TRNA SYNTHETASE, MAGNESIUM ION, TRNALEU5 UAA ISOACCEPTOR | | Authors: | Cusack, S, Palencia, A, Crepin, T, Hernandez, V, Akama, T, Baker, S.J, Bu, W, Feng, L, Freund, Y.R, Liu, L, Meewan, M, Mohan, M, Mao, W, Rock, F.L, Sexton, H, Sheoran, A, Zhang, Y, Zhang, Y, Zhou, Y, Nieman, J.A, Anugula, M.R, Keramane, E.M, Savariraj, K, Reddy, D.S, Sharma, R, Subedi, R, Singh, R, OLeary, A, Simon, N.L, DeMarsh, P.L, Mushtaq, S, Warner, M, Livermore, D.M, Alley, M.R.K, Plattner, J.J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Novel Class of Boron-Based Antibacterials with Activity Against Gram-Negative Bacteria.
Antimicrob.Agents Chemother., 57, 2013
|
|
3ZJV
 
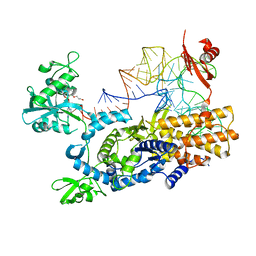 | | Ternary complex of E .coli leucyl-tRNA synthetase, tRNA(Leu) and the benzoxaborole AN3213 in the editing conformation | | Descriptor: | LEUCINE--TRNA LIGASE, TRNALEU5 UAA ISOACCEPTOR | | Authors: | Cusack, S, Palencia, A, Crepin, T, Hernandez, V, Akama, T, Baker, S.J, Bu, W, Feng, L, Freund, Y.R, Liu, L, Meewan, M, Mohan, M, Mao, W, Rock, F.L, Sexton, H, Sheoran, A, Zhang, Y, Zhang, Y, Zhou, Y, Nieman, J.A, Anugula, M.R, Keramane, E.M, Savariraj, K, Reddy, D.S, Sharma, R, Subedi, R, Singh, R, OLeary, A, Simon, N.L, DeMarsh, P.L, Mushtaq, S, Warner, M, Livermore, D.M, Alley, M.R.K, Plattner, J.J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery of a Novel Class of Boron-Based Antibacterials with Activity Against Gram-Negative Bacteria.
Antimicrob.Agents Chemother., 57, 2013
|
|
3ZJU
 
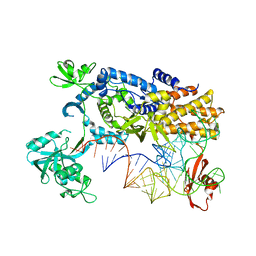 | | Ternary complex of E .coli leucyl-tRNA synthetase, tRNA(Leu) and the benzoxaborole AN3016 in the editing conformation | | Descriptor: | LEUCYL-TRNA SYNTHETASE, MAGNESIUM ION, TRNALEU5 UAA ISOACCEPTOR | | Authors: | Cusack, S, Palencia, A, Crepin, T, Hernandez, V, Akama, T, Baker, S.J, Bu, W, Feng, L, Freund, Y.R, Liu, L, Meewan, M, Mohan, M, Mao, W, Rock, F.L, Sexton, H, Sheoran, A, Zhang, Y, Zhang, Y, Zhou, Y, Nieman, J.A, Anugula, M.R, Keramane, E.M, Savariraj, K, Reddy, D.S, Sharma, R, Subedi, R, Singh, R, OLeary, A, Simon, N.L, DeMarsh, P.L, Mushtaq, S, Warner, M, Livermore, D.M, Alley, M.R.K, Plattner, J.J. | | Deposit date: | 2013-01-18 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a Novel Class of Boron-Based Antibacterials with Activity Against Gram-Negative Bacteria.
Antimicrob.Agents Chemother., 57, 2013
|
|
8WT1
 
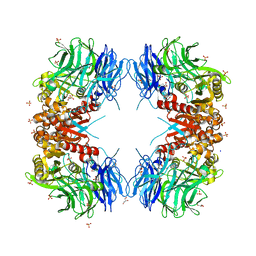 | | Crystal structure of S9 carboxypeptidase from Geobacillus sterothermophilus | | Descriptor: | ALANINE, CITRATE ANION, GLYCEROL, ... | | Authors: | Chandravanshi, K, Kumar, A, Sen, C, Singh, R, Bhange, G.B, Makde, R.D. | | Deposit date: | 2023-10-17 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and solution scattering of Geobacillus stearothermophilus S9 peptidase reveal structural adaptations for carboxypeptidase activity.
Febs Lett., 598, 2024
|
|
1EE1
 
 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS COMPLEXED WITH ONE MOLECULE ATP, TWO MOLECULES DEAMIDO-NAD+ AND ONE MG2+ ION | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NH(3)-DEPENDENT NAD(+) SYNTHETASE, ... | | Authors: | Devedjiev, Y, Symersky, J, Singh, R, Jedrzejas, M, Brouillette, C, Brouillette, W, Muccio, D, Chattopadhyay, D, Delucas, L. | | Deposit date: | 2000-01-28 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
6ZGQ
 
 | | AceL NrdHF class 3 split intein GSH linked splice inactive variant - C124A, N146A | | Descriptor: | AceL NrdHF-1-1 Intein, IODIDE ION | | Authors: | Hoffmann, S, Mootz, H.D, Kuemmel, D, Singh, R. | | Deposit date: | 2020-06-19 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and Structural Characterization of an Unusual and Naturally Split Class 3 Intein.
Chembiochem, 22, 2021
|
|
1IH8
 
 | | NH3-dependent NAD+ Synthetase from Bacillus subtilis Complexed with AMP-CPP and Mg2+ ions. | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, NH(3)-DEPENDENT NAD(+) synthetase | | Authors: | Devedjiev, Y, Symersky, J, Singh, R, Jedrzejas, M, Brouillette, C, Brouillette, W, Muccio, D, Chattopadhyay, D, DeLucas, L. | | Deposit date: | 2001-04-18 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1IFX
 
 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS COMPLEXED WITH TWO MOLECULES DEAMIDO-NAD | | Descriptor: | NH(3)-DEPENDENT NAD(+) SYNTHETASE, NICOTINIC ACID ADENINE DINUCLEOTIDE | | Authors: | Devedjiev, Y, Symersky, J, Singh, R, Brouillette, W, Muccio, D, Jedrzejas, M, Brouillette, C, DeLucas, L. | | Deposit date: | 2001-04-13 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1FYD
 
 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS COMPLEXED WITH ONE MOLECULE AMP, ONE PYROPHOSPHATE ION AND ONE MG2+ ION | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, NH(3)-DEPENDENT NAD(+) SYNTHETASE, ... | | Authors: | Devedjiev, Y, Symersky, J, Singh, R, Brouillette, W, Muccio, D, Jedrzejas, M, Brouillette, C, DeLucas, L. | | Deposit date: | 2000-09-28 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1MU6
 
 | | Crystal Structure of Thrombin in Complex with L-378,622 | | Descriptor: | 2-(6-CHLORO-3-{[2,2-DIFLUORO-2-(2-PYRIDINYL)ETHYL]AMINO}-2-OXO-1(2H)-PYRAZINYL)-N-[(2-FLUORO-6-PYRIDINYL)METHYL]ACETAMIDE, HIRUDIN IIB, THROMBIN | | Authors: | Burgey, C.S, Robinson, K.A, Lyle, T.A, Sanderson, P.E, Lewis, S.D, Lucas, B.J, Krueger, J.A, Singh, R, Miller-Stein, C, White, R.B, Wong, B, Lyle, E.A, Williams, P.D, Coburn, C.A, Dorsey, B.D, Barrow, J.C, Stranieri, M.T, Holahan, M.A, Sitko, G.R, Cook, J.J, McMasters, D.R, McDonough, C.M, Sanders, W.M, Wallace, A.A, Clayton, F.C, Bohn, D, Leonard, Y.M, Detwiler Jr, T.J, Lynch Jr, J.J, Yan, Y, Chen, Z, Kuo, L, Gardell, S.J, Shafer, J.A, Vacca, J.P.J. | | Deposit date: | 2002-09-23 | | Release date: | 2004-04-06 | | Last modified: | 2021-07-21 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Metabolism-directed optimization of 3-aminopyrazinone acetamide thrombin inhibitors. Development of an orally bioavailable series containing P1 and P3 pyridines.
J.Med.Chem., 46, 2003
|
|
1MU8
 
 | | thrombin-hirugen_l-378,650 | | Descriptor: | 2-(6-CHLORO-3-{[2,2-DIFLUORO-2-(2-PYRIDINYL)ETHYL]AMINO}-2-OXO-1(2H)-PYRAZINYL)-N-[(2-FLUORO-3-METHYL-6-PYRIDINYL)METHYL]ACETAMIDE, HIRUDIN IIB, THROMBIN | | Authors: | Burgey, C.S, Robinson, K.A, Lyle, T.A, Sanderson, P.E, Lewis, S.D, Lucas, B.J, Krueger, J.A, Singh, R, Miller-Stein, C, White, R.B, Wong, B, Lyle, E.A, Williams, P.D, Coburn, C.A, Dorsey, B.D, Barrow, J.C, Stranieri, M.T, Holahan, M.A, Sitko, G.R, Cook, J.J, McMasters, D.R, McDonough, C.M, Sanders, W.M, Wallace, A.A, Clayton, F.C, Bohn, D, Leonard, Y.M, Detwiler Jr, T.J, Lynch Jr, J.J, Yan, Y, Chen, Z, Kuo, L, Gardell, S.J, Shafer, J.A, Vacca, J.P.J. | | Deposit date: | 2002-09-23 | | Release date: | 2004-04-06 | | Last modified: | 2021-07-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metabolism-directed optimization of 3-aminopyrazinone acetamide thrombin inhibitors. Development of an orally bioavailable series containing P1 and P3 pyridines.
J.Med.Chem., 46, 2003
|
|
2BV3
 
 | | Crystal structure of a mutant elongation factor G trapped with a GTP analogue | | Descriptor: | ELONGATION FACTOR G, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Hansson, S, Singh, R, Gudkov, A.T, Liljas, A, Logan, D.T. | | Deposit date: | 2005-06-22 | | Release date: | 2005-08-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Mutant Elongation Factor G Trapped with a GTP Analogue.
FEBS Lett., 579, 2005
|
|
