4BXI
 
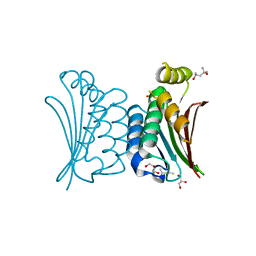 | | Crystal structure of ATP binding domain of AgrC from Staphylococcus aureus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACCESSORY GENE REGULATOR PROTEIN C, ACETATE ION, ... | | Authors: | Srivastava, S.K, Rajasree, K, Gopal, B. | | Deposit date: | 2013-07-12 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Influence of the Agrc-Agra Complex in the Response Time of Staphylococcus Aureus Quorum Sensing
J.Bacteriol., 196, 2014
|
|
1ZAU
 
 | |
3Q86
 
 | |
3Q83
 
 | |
3Q8Y
 
 | |
3Q89
 
 | |
3Q8U
 
 | |
3Q8V
 
 | |
3PQ1
 
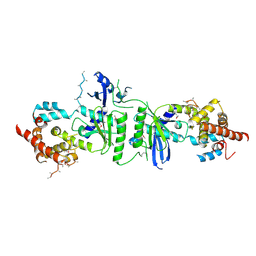 | | Crystal structure of human mitochondrial poly(A) polymerase (PAPD1) | | Descriptor: | Poly(A) RNA polymerase | | Authors: | Bai, Y, Srivastava, S.K, Chang, J.H, Tong, L. | | Deposit date: | 2010-11-25 | | Release date: | 2011-03-30 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for dimerization and activity of human PAPD1, a noncanonical poly(A) polymerase.
Mol.Cell, 41, 2011
|
|
2F2K
 
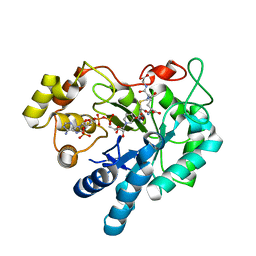 | | Aldose reductase tertiary complex with NADPH and DEG | | Descriptor: | Aldose reductase, GAMMA-GLUTAMYL-S-(1,2-DICARBOXYETHYL)CYSTEINYLGLYCINE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Singh, R, White, M.A, Ramana, K.V, Petrash, J.M, Watowich, S.J, Bhatnagar, A, Srivastava, S.K. | | Deposit date: | 2005-11-17 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure of a glutathione conjugate bound to the active site of aldose reductase.
Proteins, 64, 2006
|
|
