2RT9
 
 | | Solution structure of a regulatory domain of meiosis inhibitor | | Descriptor: | F-box only protein 43, ZINC ION | | Authors: | Shoji, S, Muto, Y, Ikeda, M, He, F, Tsuda, K, Ohsawa, N, Akasaka, R, Terada, T, Wakiyama, M, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-05 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The zinc-binding region (ZBR) fragment of Emi2 can inhibit APC/C by targeting its association with the coactivator Cdc20 and UBE2C-mediated ubiquitylation
FEBS Open Bio, 4, 2014
|
|
2RQ4
 
 | | Refinement of RNA binding domain 3 in CUG triplet repeat RNA-binding protein 1 | | Descriptor: | CUG-BP- and ETR-3-like factor 1 | | Authors: | Tsuda, K, Kuwasako, K, Takahashi, M, Someya, T, Inoue, M, Terada, T, Kobayashi, N, Shirouzu, M, Kigawa, T, Guntert, P, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-01-19 | | Release date: | 2009-08-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3
Nucleic Acids Res., 2009
|
|
2ROK
 
 | | Solution structure of the cap-binding domain of PARN complexed with the cap analog | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, poly(A)-specific ribonuclease | | Authors: | Nagata, T, Suzuki, S, Endo, R, Shirouzu, M, Terada, T, Inoue, M, Kigawa, T, Guntert, P, Hayashizaki, Y, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-03-28 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The RRM domain of poly(A)-specific ribonuclease has a noncanonical binding site for mRNA cap analog recognition.
Nucleic Acids Res., 36, 2008
|
|
2RQC
 
 | | Solution Structure of RNA-binding domain 3 of CUGBP1 in complex with RNA (UG)3 | | Descriptor: | 5'-R(*UP*GP*UP*GP*UP*G)-3', CUG-BP- and ETR-3-like factor 1 | | Authors: | Tsuda, K, Kuwasako, K, Takahashi, M, Someya, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-04-09 | | Release date: | 2009-08-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3
Nucleic Acids Res., 37, 2009
|
|
2RRB
 
 | | Refinement of RNA binding domain in human Tra2 beta protein | | Descriptor: | cDNA FLJ40872 fis, clone TUTER2000283, highly similar to Homo sapiens transformer-2-beta (SFRS10) gene | | Authors: | Tsuda, K, Kuwasako, K, Takahashi, M, Someya, T, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Sugano, S, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-06-17 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the dual RNA-recognition modes of human Tra2-beta RRM.
Nucleic Acids Res., 39, 2011
|
|
2RU3
 
 | | Solution structure of c.elegans SUP-12 RRM in complex with RNA | | Descriptor: | Protein SUP-12, isoform a, RNA (5'-R(*GP*UP*GP*UP*GP*C)-3') | | Authors: | Takahashi, M, Kuwasako, K, Unzai, S, Tsuda, K, Yoshikawa, S, He, F, Kobayashi, N, Guntert, P, Shirouzu, M, Ito, T, Tanaka, A, Yokoyama, S, Hagiwara, M, Kuroyanagi, H, Muto, Y. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | RBFOX and SUP-12 sandwich a G base to cooperatively regulate tissue-specific splicing
Nat.Struct.Mol.Biol., 21, 2014
|
|
4LM5
 
 | | Crystal structure of Pim1 in complex with 2-{4-[(3-aminopropyl)amino]quinazolin-2-yl}phenol (resulting from displacement of SKF86002) | | Descriptor: | 2-{4-[(3-aminopropyl)amino]quinazolin-2-yl}phenol, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-10 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LMU
 
 | | Crystal structure of Pim1 in complex with the inhibitor Quercetin (resulting from displacement of SKF86002) | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-11 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LL5
 
 | | Crystal Structure of Pim-1 in complex with the fluorescent compound SKF86002 | | Descriptor: | 6-(4-fluorophenyl)-5-(pyridin-4-yl)-2,3-dihydroimidazo[2,1-b][1,3]thiazole, CALCIUM ION, GLYCEROL, ... | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2RUG
 
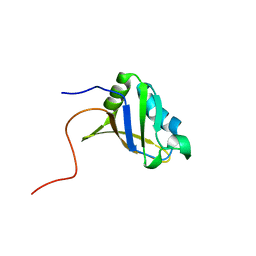 | | Refined solution structure of the first RNA recognition motif domain in CPEB3 | | Descriptor: | Cytoplasmic polyadenylation element-binding protein 3 | | Authors: | Tsuda, K, Kuwasako, K, Nagata, T, Takahashi, M, Kigawa, T, Kobayashi, N, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2014-04-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Novel RNA recognition motif domain in the cytoplasmic polyadenylation element binding protein 3.
Proteins, 82, 2014
|
|
2RS7
 
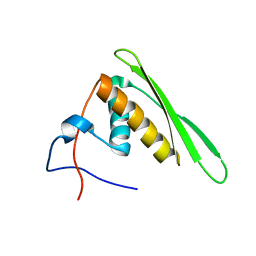 | | Solution structure of the second dsRBD from RNA helicase A | | Descriptor: | ATP-dependent RNA helicase A | | Authors: | Nagata, T, Muto, Y, Tsuda, K, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-11-29 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the double-stranded RNA-binding domains from RNA helicase A
Proteins, 80, 2012
|
|
2RS6
 
 | | Solution structure of the N-terminal dsRBD from RNA helicase A | | Descriptor: | ATP-dependent RNA helicase A | | Authors: | Nagata, T, Muto, Y, Tsuda, K, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-11-29 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the double-stranded RNA-binding domains from RNA helicase A
Proteins, 80, 2012
|
|
2PA2
 
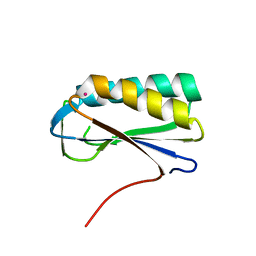 | | Crystal structure of human Ribosomal protein L10 core domain | | Descriptor: | 60S ribosomal protein L10, POTASSIUM ION | | Authors: | Nishimura, M, Kaminishi, T, Takemoto, C, Kawazoe, M, Yoshida, T, Tanaka, A, Sugano, S, Shirouzu, M, Ohkubo, T, Yokoyama, S, Kobayashi, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Human Ribosomal Protein L10 Core Domain Reveals Eukaryote-Specific Motifs in Addition to the Conserved Fold
J.Mol.Biol., 377, 2008
|
|
2PSM
 
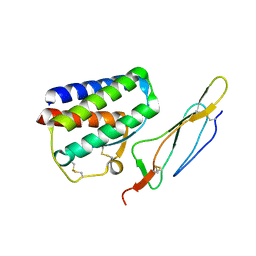 | | Crystal structure of Interleukin 15 in complex with Interleukin 15 receptor alpha | | Descriptor: | BENZAMIDINE, Interleukin-15, Interleukin-15 receptor alpha chain | | Authors: | Olsen, S.K, Murayama, K, Kishishita, S, Kukimoto-Niino, M, Terada, T, Shirouzu, M, Ota, N, Kanagawa, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of the Interleukin-15{middle dot}Interleukin-15 Receptor {alpha} Complex: INSIGHTS INTO TRANS AND CIS PRESENTATION
J.Biol.Chem., 282, 2007
|
|
2RNE
 
 | | Solution structure of the second RNA recognition motif (RRM) of TIA-1 | | Descriptor: | Tia1 protein | | Authors: | Takahashi, M, Kuwasako, K, Abe, C, Tsuda, K, Inoue, M, Terada, T, Shirouzu, M, Kobayashi, N, Kigawa, T, Taguchi, S, Guntert, P, Hayashizaki, Y, Tanaka, A, Muto, Y, Yokoyama, S. | | Deposit date: | 2007-12-19 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second RNA recognition motif (RRM) domain of murine T cell intracellular antigen-1 (TIA-1) and its RNA recognition mode
Biochemistry, 47, 2008
|
|
2RNL
 
 | | Solution structure of the EGF-like domain from human Amphiregulin | | Descriptor: | Amphiregulin | | Authors: | Qin, X, Hayashi, F, Terada, T, Shirouzu, M, Watanabe, S, Kigawa, T, Yabuta, N, Nojima, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-01-11 | | Release date: | 2009-01-20 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the EGF-like domain from human Amphiregulin
To be Published
|
|
6KS1
 
 | | Crystal structure of the human adiponectin receptor 2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, ... | | Authors: | Tanabe, H, Fujii, Y, Nakamura, Y, Hosaka, T, Okada-Iwabu, M, Iwabu, M, Kimura-Someya, T, Shirouzu, M, Yamauchi, T, Kadowaki, T, Yokoyama, S. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human adiponectin receptor AdipoR1 assumes closed and open structures.
Commun Biol, 3, 2020
|
|
1EF5
 
 | | SOLUTION STRUCTURE OF THE RAS-BINDING DOMAIN OF RGL | | Descriptor: | RGL | | Authors: | Kigawa, T, Endo, M, Ito, Y, Shirouzu, M, Kikuchi, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-02-07 | | Release date: | 2000-02-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Ras-binding domain of RGL.
FEBS Lett., 441, 1998
|
|
6KS0
 
 | | Crystal structure of the human adiponectin receptor 1 | | Descriptor: | Adiponectin receptor protein 1, PHOSPHATE ION, The heavy chain variable domain (Antibody), ... | | Authors: | Tanabe, H, Fujii, Y, Nakamura, Y, Hosaka, T, Okada-Iwabu, M, Iwabu, M, Kimura-Someya, T, Shirouzu, M, Yamauchi, T, Kadowaki, T, Yokoyama, S. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Human adiponectin receptor AdipoR1 assumes closed and open structures.
Commun Biol, 3, 2020
|
|
6KRO
 
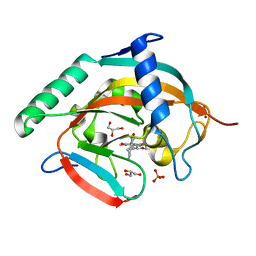 | | Tankyrase-2 in complex with RK-582 | | Descriptor: | 6-[(2S,6R)-2,6-dimethylmorpholin-4-yl]-4-fluoranyl-1-methyl-1'-(8-methyl-4-oxidanylidene-3,5,6,7-tetrahydropyrido[2,3-d]pyrimidin-2-yl)spiro[indole-3,4'-piperidine]-2-one, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Tsumura, T, Okue, M, Shirouzu, M, Seimiya, H, Umehara, T. | | Deposit date: | 2019-08-22 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and Discovery of an Orally Efficacious Spiroindolinone-Based Tankyrase Inhibitor for the Treatment of Colon Cancer.
J.Med.Chem., 63, 2020
|
|
6LPK
 
 | | A2AR crystallized in EROCOC17+4, LCP-SFX at 293 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
6LM0
 
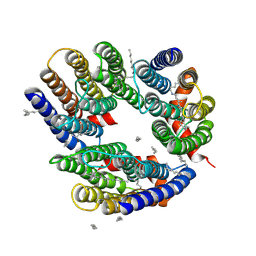 | | The crystal structure of cyanorhodopsin (CyR) N2098R from cyanobacteria Calothrix sp. NIES-2098 | | Descriptor: | DECANE, HEXANE, N-OCTANE, ... | | Authors: | Hosaka, T, Kimura-Someya, T, Shirouzu, M. | | Deposit date: | 2019-12-24 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A unique clade of light-driven proton-pumping rhodopsins evolved in the cyanobacterial lineage.
Sci Rep, 10, 2020
|
|
6M04
 
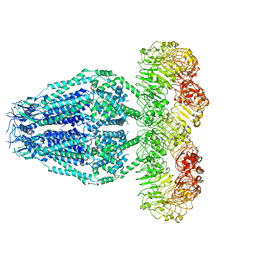 | | Structure of the human homo-hexameric LRRC8D channel at 4.36 Angstroms | | Descriptor: | Volume-regulated anion channel subunit LRRC8D | | Authors: | Nakamura, R, Kasuya, G, Yokoyama, T, Shirouzu, M, Ishitani, R, Nureki, O. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Cryo-EM structure of the volume-regulated anion channel LRRC8D isoform identifies features important for substrate permeation.
Commun Biol, 3, 2020
|
|
6LPJ
 
 | | A2AR crystallized in EROCOC17+4, LCP-SFX at 277 K | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Ihara, K, Hato, M, Nakane, T, Yamashita, K, Kimura-Someya, T, Hosaka, T, Ishizuka-Katsura, Y, Tanaka, R, Tanaka, T, Sugahara, M, Hirata, K, Yamamoto, M, Nureki, O, Tono, K, Nango, E, Iwata, S, Shirouzu, M. | | Deposit date: | 2020-01-10 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Isoprenoid-chained lipid EROCOC 17+4 : a new matrix for membrane protein crystallization and a crystal delivery medium in serial femtosecond crystallography.
Sci Rep, 10, 2020
|
|
6LM1
 
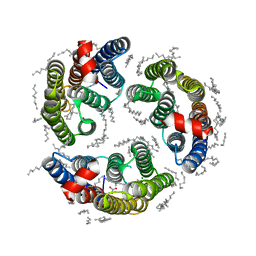 | | The crystal structure of cyanorhodopsin (CyR) N4075R from cyanobacteria Tolypothrix sp. NIES-4075 | | Descriptor: | DECANE, DODECANE, HEXADECANE, ... | | Authors: | Hosaka, T, Kimura-Someya, T, Shirouzu, M. | | Deposit date: | 2019-12-24 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A unique clade of light-driven proton-pumping rhodopsins evolved in the cyanobacterial lineage.
Sci Rep, 10, 2020
|
|
