3VIR
 
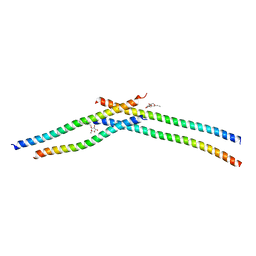 | | Crystal strcture of Swi5 from fission yeast | | 分子名称: | Mating-type switching protein swi5, octyl beta-D-glucopyranoside | | 著者 | Kuwabara, N, Yamada, N, Hashimoto, H, Sato, M, Iwasaki, H, Shimizu, T. | | 登録日 | 2011-10-06 | | 公開日 | 2012-08-22 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Mechanistic insights into the activation of Rad51-mediated strand exchange from the structure of a recombination activator, the Swi5-Sfr1 complex
Structure, 20, 2012
|
|
3VLA
 
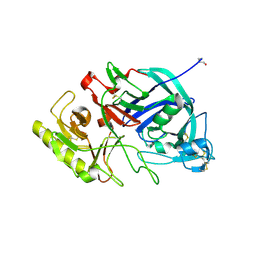 | | Crystal structure of edgp | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EDGP | | 著者 | Yoshizawa, T, Shimizu, T, Hirano, H, Sato, M, Hashimoto, H. | | 登録日 | 2011-11-30 | | 公開日 | 2012-04-18 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (0.95 Å) | | 主引用文献 | Structural basis for inhibition of xyloglucan-specific endo-beta-1,4-glucanase (XEG) by XEG-protein inhibitor
J.Biol.Chem., 287, 2012
|
|
4TKC
 
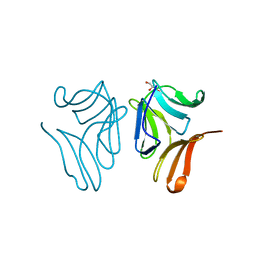 | | Japanese Marasmius oreades lectin complexed with mannose | | 分子名称: | GLYCEROL, Mannose recognizing lectin, alpha-D-mannopyranose, ... | | 著者 | Noma, Y, Shimokawa, M, Maeganeku, C, Motoshima, H, Watanabe, K, Minami, Y, Yagi, F. | | 登録日 | 2014-05-26 | | 公開日 | 2015-06-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Structure of Japanese Marasmius oreades lectin complexed with mannose.
To Be Published
|
|
6IGX
 
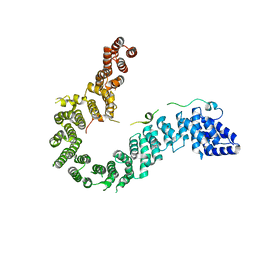 | | Crystal structure of human CAP-G in complex with CAP-H | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Condensin complex subunit 2, Condensin complex subunit 3 | | 著者 | Hara, K, Migita, T, Shimizu, K, Hashimoto, H. | | 登録日 | 2018-09-26 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.995 Å) | | 主引用文献 | Structural basis of HEAT-kleisin interactions in the human condensin I subcomplex.
Embo Rep., 20, 2019
|
|
5UZI
 
 | | Insights into Watson-Crick/Hoogsteen Breathing Dynamics and Damage Repair from the Solution Structure and Dynamic Ensemble of DNA Duplexes containing m1A - A6-DNAm1A16 structure | | 分子名称: | DNA (5'-D(*CP*GP*AP*TP*TP*TP*TP*TP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*(M1A)P*AP*AP*AP*AP*AP*TP*CP*G)-3') | | 著者 | Sathyamoorthy, B, Shi, H, Xue, Y, Al-Hashimi, H.M. | | 登録日 | 2017-02-26 | | 公開日 | 2017-04-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Insights into Watson-Crick/Hoogsteen breathing dynamics and damage repair from the solution structure and dynamic ensemble of DNA duplexes containing m1A.
Nucleic Acids Res., 45, 2017
|
|
5H6M
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1 | | 分子名称: | 1,2-ETHANEDIOL, Pierisin-1 | | 著者 | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | 登録日 | 2016-11-14 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5H6N
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1, autoinhibitory form | | 分子名称: | Pierisin-1 | | 著者 | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | 登録日 | 2016-11-14 | | 公開日 | 2017-08-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5H6K
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1 | | 分子名称: | 1,2-ETHANEDIOL, Pierisin-1 | | 著者 | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | 登録日 | 2016-11-14 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5H6L
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1 in complex with beta-NAD+ | | 分子名称: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pierisin-1 | | 著者 | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | 登録日 | 2016-11-14 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
6IRW
 
 | | Crystal structure of the human cap-specific adenosine methyltransferase bound to SAH | | 分子名称: | Phosphorylated CTD-interacting factor 1, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | 登録日 | 2018-11-14 | | 公開日 | 2018-12-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
5H6J
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1 in complex with beta-NAD+ | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pierisin-1 | | 著者 | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | 登録日 | 2016-11-14 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
6IRX
 
 | |
6IRY
 
 | | Crystal structure of the zebrafish cap-specific adenosine methyltransferase bound to SAH | | 分子名称: | 1,2-ETHANEDIOL, PDX1 C-terminal-inhibiting factor 1, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | 登録日 | 2018-11-14 | | 公開日 | 2018-12-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
8RXR
 
 | | Crystal structure of VPS34 in complex with inhibitor SB02024 | | 分子名称: | 4-[(3R)-3-methylmorpholin-4-yl]-2-[(2R)-2-(trifluoromethyl)piperidin-1-yl]-3H-pyridin-6-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | 著者 | Tresaugues, L, Yu, Y, Bogdan, M, Parpal, S, Silvander, C, Lindstrom, J, Simeon, J, Timson, M.J, Al-Hashimi, H, Smith, B.D, Flynn, D.L, Viklund, J, Martinsson, J, De Milito, A, Andersson, M. | | 登録日 | 2024-02-07 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Combining VPS34 inhibitors with STING agonists enhances type I interferon signaling and anti-tumor efficacy.
Mol Oncol, 2024
|
|
3VU7
 
 | | Crystal structure of REV1-REV7-REV3 ternary complex | | 分子名称: | DNA polymerase zeta catalytic subunit, DNA repair protein REV1, Mitotic spindle assembly checkpoint protein MAD2B | | 著者 | Kikuchi, S, Hara, K, Shimizu, T, Sato, M, Hashimoto, H. | | 登録日 | 2012-06-20 | | 公開日 | 2012-08-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis of recruitment of DNA polymerase [zeta] by interaction between REV1 and REV7 proteins
J.Biol.Chem., 287, 2012
|
|
3VJI
 
 | | Human PPAR gamma ligand binding domain in complex with JKPL53 | | 分子名称: | (2S)-2-{4-butoxy-3-[({4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]benzoyl}amino)methyl]benzyl}butanoic acid, Peroxisome proliferator-activated receptor gamma | | 著者 | Tomioka, D, Kuwabara, N, Hashimoto, H, Sato, M, Shimizu, T. | | 登録日 | 2011-10-20 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Peroxisome proliferator-activated receptors (PPARs) have multiple binding points that accommodate ligands in various conformations: phenylpropanoic acid-type PPAR ligands bind to PPAR in different conformations, depending on the subtype.
J.Med.Chem., 55, 2012
|
|
3VJH
 
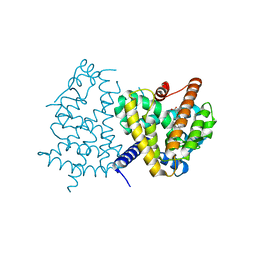 | | Human PPAR GAMMA ligand binding domain in complex with JKPL35 | | 分子名称: | (2S)-2-[4-methoxy-3-({[4-(trifluoromethyl)benzoyl]amino}methyl)benzyl]pentanoic acid, Peroxisome proliferator-activated receptor gamma | | 著者 | Tomioka, D, Kuwabara, N, Hashimoto, H, Sato, M, Shimizu, T. | | 登録日 | 2011-10-20 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Peroxisome proliferator-activated receptors (PPARs) have multiple binding points that accommodate ligands in various conformations: phenylpropanoic acid-type PPAR ligands bind to PPAR in different conformations, depending on the subtype.
J.Med.Chem., 55, 2012
|
|
8Y2S
 
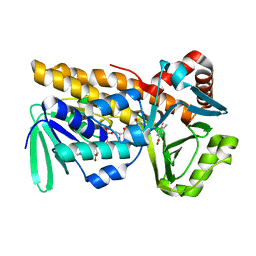 | | P-hydroxybenzoate hydroxylase complexed with 4-hydroxy-3-methylbenzoic acid | | 分子名称: | 3-methyl-4-oxidanyl-benzoic acid, 4-hydroxybenzoate 3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Hara, K, Hashimoto, H, Matsushita, T, Kishimoto, S, Watanabe, K. | | 登録日 | 2024-01-27 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Functional Enhancement of Flavin-Containing Monooxygenase through Machine Learning Methodology
Acs Catalysis, 14, 2024
|
|
1GE9
 
 | | SOLUTION STRUCTURE OF THE RIBOSOME RECYCLING FACTOR | | 分子名称: | RIBOSOME RECYCLING FACTOR | | 著者 | Yoshida, T, Uchiyama, S, Nakano, H, Kashimori, H, Kijima, H, Ohshima, T, Saihara, Y, Ishino, T, Shimahara, T, Yoshida, T, Yokose, K, Ohkubo, T, Kaji, A, Kobayashi, Y. | | 登録日 | 2000-10-19 | | 公開日 | 2001-05-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the ribosome recycling factor from Aquifex aeolicus.
Biochemistry, 40, 2001
|
|
2CT9
 
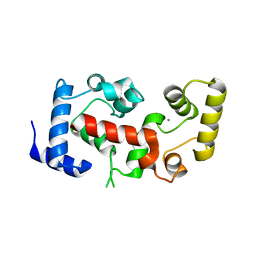 | | The crystal structure of calcineurin B homologous proein 1 (CHP1) | | 分子名称: | CALCIUM ION, Calcium-binding protein p22 | | 著者 | Naoe, Y, Arita, K, Hashimoto, H, Kanazawa, H, Sato, M, Shimizu, T. | | 登録日 | 2005-05-23 | | 公開日 | 2005-07-05 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural characterization of calcineurin B homologous protein 1
J.Biol.Chem., 280, 2005
|
|
2DCT
 
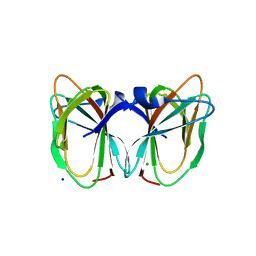 | | Crystal structure of the TT1209 from Thermus thermophilus HB8 | | 分子名称: | CHLORIDE ION, SODIUM ION, hypothetical protein TTHA0104 | | 著者 | Asada, Y, Sugahara, M, Shimizu, K, Yamamoto, H, Shimada, H, Nakamoto, T, Ono, N, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-01-12 | | 公開日 | 2006-01-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Crystal structure of the TT1209 from Thermus thermophilus HB8
To be Published
|
|
3J3Z
 
 | | Structure of MA28-7 neutralizing antibody Fab fragment from electron cryo-microscopy of enterovirus 71 complexed with a Fab fragment | | 分子名称: | MA28-7 neutralizing antibody heavy chain, MA28-7 neutralizing antibody light chain | | 著者 | Lee, H, Cifuente, J.O, Ashley, R.E, Conway, J.F, Makhov, A.M, Tano, Y, Shimizu, H, Nishimura, Y, Hafenstein, S. | | 登録日 | 2013-05-21 | | 公開日 | 2013-08-28 | | 最終更新日 | 2018-07-18 | | 実験手法 | ELECTRON MICROSCOPY (23.4 Å) | | 主引用文献 | A strain-specific epitope of enterovirus 71 identified by cryo-electron microscopy of the complex with fab from neutralizing antibody.
J.Virol., 87, 2013
|
|
8WT9
 
 | | Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the post-strand exchange state (Holliday junction resolution) | | 分子名称: | IS621 transposase, MAGNESIUM ION, bridge RNA, ... | | 著者 | Hiraizumi, M, Yamashita, K, Nishimasu, H. | | 登録日 | 2023-10-18 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structural mechanism of bridge RNA-guided recombination.
Nature, 630, 2024
|
|
8WT7
 
 | | Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the pre-strand exchange locked state | | 分子名称: | IS621 transposase, MAGNESIUM ION, bridge RNA, ... | | 著者 | Hiraizumi, M, Yamashita, K, Nishimasu, H. | | 登録日 | 2023-10-18 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structural mechanism of bridge RNA-guided recombination.
Nature, 630, 2024
|
|
8WT8
 
 | | Cryo-EM structure of the IS621 recombinase in complex with bridge RNA, donor DNA, and target DNA in the post-strand exchange state (Holliday junction intermediate) | | 分子名称: | IS621 transposase, MAGNESIUM ION, bridge RNA, ... | | 著者 | Hiraizumi, M, Yamashita, K, Nishimasu, H. | | 登録日 | 2023-10-18 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural mechanism of bridge RNA-guided recombination.
Nature, 630, 2024
|
|
