4DNW
 
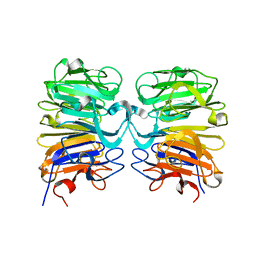 | | Crystal structure of UVB-resistance protein UVR8 | | 分子名称: | AT5g63860/MGI19_6 | | 著者 | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Wang, J, Shi, Y. | | 登録日 | 2012-02-09 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.773 Å) | | 主引用文献 | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
4F23
 
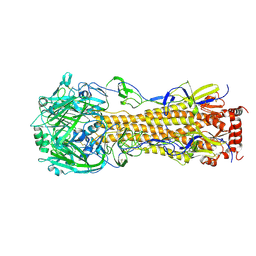 | | Influenza A virus hemagglutinin H16 HA0 structure with an alpha-helix conformation in the cleavage site: a potential drug target | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | 著者 | Lu, X, Shi, Y, Gao, F, Xiao, H, Qi, J, Gao, G.F. | | 登録日 | 2012-05-07 | | 公開日 | 2012-11-21 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Insights into Avian Influenza Virus Pathogenicity: the Hemagglutinin Precursor HA0 of Subtype H16 Has an Alpha-Helix Structure in Its Cleavage Site with Inefficient HA1/HA2 Cleavage.
J.Virol., 86, 2012
|
|
4GND
 
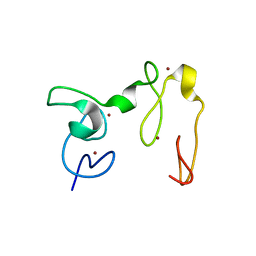 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains | | 分子名称: | Histone-lysine N-methyltransferase NSD3, ZINC ION | | 著者 | Li, F, He, C, Wu, J, Shi, Y. | | 登録日 | 2012-08-17 | | 公開日 | 2013-01-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
4GNE
 
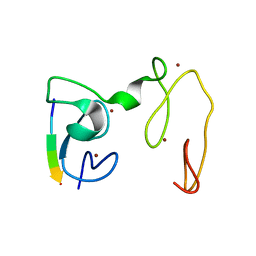 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3 peptide 1-7 | | 分子名称: | Histone H3.3, Histone-lysine N-methyltransferase NSD3, ZINC ION | | 著者 | Li, F, He, C, Wu, J, Shi, Y. | | 登録日 | 2012-08-17 | | 公開日 | 2013-01-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
5YXA
 
 | | Crystal structure of the C-terminal fragment of NS1 protein from yellow fever virus | | 分子名称: | Non-structural protein 1 | | 著者 | Wang, H, Song, H, Qi, J, Shi, Y, Gao, G.F. | | 登録日 | 2017-12-04 | | 公開日 | 2018-01-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of the C-terminal fragment of NS1 protein from yellow fever virus.
Sci China Life Sci, 60, 2017
|
|
4GNF
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3 peptide 1-15 | | 分子名称: | Histone H3.3, Histone-lysine N-methyltransferase NSD3, ZINC ION | | 著者 | Li, F, He, C, Wu, J, Shi, Y. | | 登録日 | 2012-08-17 | | 公開日 | 2013-01-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
4GNG
 
 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3K9me3 peptide | | 分子名称: | GLYCEROL, Histone H3.3, Histone-lysine N-methyltransferase NSD3, ... | | 著者 | Li, F, He, C, Wu, J, Shi, Y. | | 登録日 | 2012-08-17 | | 公開日 | 2013-01-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
4GXL
 
 | | The crystal structure of Galectin-8 C-CRD in complex with NDP52 | | 分子名称: | GLYCEROL, Galectin-8, Peptide from Calcium-binding and coiled-coil domain-containing protein 2 | | 著者 | Li, S, Wandel, M.P, Li, F, Liu, Z, He, C, Wu, J, Shi, Y, Randow, F. | | 登録日 | 2012-09-04 | | 公開日 | 2013-05-08 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.023 Å) | | 主引用文献 | Sterical hindrance promotes selectivity of the autophagy cargo receptor NDP52 for the danger receptor galectin-8 in antibacterial autophagy
Sci.Signal., 6, 2013
|
|
4FIU
 
 | | The structure of hemagglutinin of H16 subtype influenza virus with V327G mutation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, hemagglutinin | | 著者 | Lu, X, Shi, Y, Gao, F, Xiao, H, Wang, M, Qi, J, Gao, G.F. | | 登録日 | 2012-06-11 | | 公開日 | 2012-11-21 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | Insights into Avian Influenza Virus Pathogenicity: the Hemagglutinin Precursor HA0 of Subtype H16 Has an Alpha-Helix Structure in Its Cleavage Site with Inefficient HA1/HA2 Cleavage.
J.Virol., 86, 2012
|
|
1LCY
 
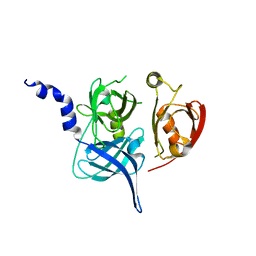 | | Crystal Structure of the Mitochondrial Serine Protease HtrA2 | | 分子名称: | HtrA2 serine protease | | 著者 | Li, W, Srinivasula, S.M, Chai, J, Li, P, Wu, J.W, Zhang, Z, Alnemri, E.S, Shi, Y. | | 登録日 | 2002-04-07 | | 公開日 | 2002-05-22 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural insights into the pro-apoptotic function of mitochondrial serine protease HtrA2/Omi.
Nat.Struct.Biol., 9, 2002
|
|
5YZG
 
 | | The Cryo-EM Structure of Human Catalytic Step I Spliceosome (C complex) at 4.1 angstrom resolution | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Zhan, X, Yan, C, Zhang, X, Lei, J, Shi, Y. | | 登録日 | 2017-12-14 | | 公開日 | 2018-08-08 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structure of a human catalytic step I spliceosome
Science, 359, 2018
|
|
4DNV
 
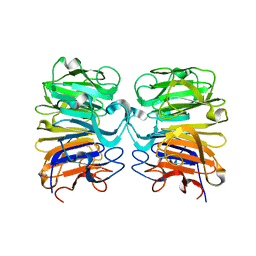 | | Crystal structure of the W285F mutant of UVB-resistance protein UVR8 | | 分子名称: | AT5g63860/MGI19_6 | | 著者 | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | 登録日 | 2012-02-09 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
5ZWN
 
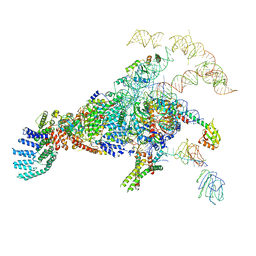 | | Cryo-EM structure of the yeast pre-B complex at an average resolution of 3.3 angstrom (Part II: U1 snRNP region) | | 分子名称: | 56 kDa U1 small nuclear ribonucleoprotein component, Pre-mRNA-processing factor 39, Pre-mRNA-splicing ATP-dependent RNA helicase PRP28, ... | | 著者 | Bai, R, Wan, R, Yan, C, Lei, J, Shi, Y. | | 登録日 | 2018-05-16 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures of the fully assembledSaccharomyces cerevisiaespliceosome before activation
Science, 360, 2018
|
|
4HYD
 
 | | Structure of a presenilin family intramembrane aspartate protease in C2221 space group | | 分子名称: | Putative uncharacterized protein | | 著者 | Li, X, Dang, S, Yan, C, Wang, J, Shi, Y. | | 登録日 | 2012-11-13 | | 公開日 | 2012-12-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structure of a presenilin family intramembrane aspartate protease
Nature, 493, 2013
|
|
4HYC
 
 | | Structure of a presenilin family intramembrane aspartate protease in P2 space group | | 分子名称: | Putative uncharacterized protein | | 著者 | Li, X, Dang, S, Yan, C, Wang, J, Shi, Y. | | 登録日 | 2012-11-13 | | 公開日 | 2012-12-19 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.95 Å) | | 主引用文献 | Structure of a presenilin family intramembrane aspartate protease
Nature, 493, 2013
|
|
6J6H
 
 | | Cryo-EM structure of the yeast B*-a1 complex at an average resolution of 3.6 angstrom | | 分子名称: | ACT1 pre-mRNA, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Wan, R, Bai, R, Yan, C, Lei, J, Shi, Y. | | 登録日 | 2019-01-15 | | 公開日 | 2019-04-24 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching.
Cell, 177, 2019
|
|
5Y88
 
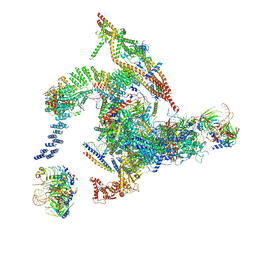 | | Cryo-EM structure of the intron-lariat spliceosome ready for disassembly from S.cerevisiae at 3.5 angstrom | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Intron lariat, ... | | 著者 | Wan, R, Yan, C, Bai, R, Lei, J, Shi, Y. | | 登録日 | 2017-08-20 | | 公開日 | 2018-08-01 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.46 Å) | | 主引用文献 | Structure of an Intron Lariat Spliceosome from Saccharomyces cerevisiae
Cell(Cambridge,Mass.), 171, 2017
|
|
6JCG
 
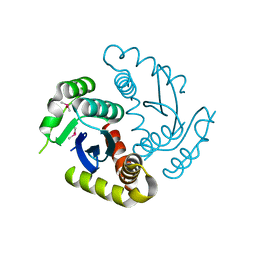 | | Room temperature structure of HIV-1 Integrase catalytic core domain by serial femtosecond crystallography. | | 分子名称: | CACODYLATE ION, Integrase | | 著者 | Park, J.H, Shi, Y, Han, J, Li, X, Kim, T.H, Yun, J.H. | | 登録日 | 2019-01-28 | | 公開日 | 2019-07-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Non-Cryogenic Structure and Dynamics of HIV-1 Integrase Catalytic Core Domain by X-ray Free-Electron Lasers.
Int J Mol Sci, 20, 2019
|
|
5Z58
 
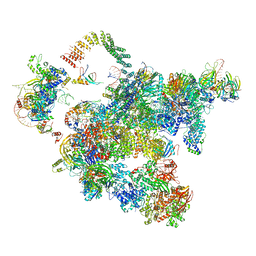 | | Cryo-EM structure of a human activated spliceosome (early Bact) at 4.9 angstrom. | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | 著者 | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | 登録日 | 2018-01-17 | | 公開日 | 2018-09-19 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
5ZWO
 
 | | Cryo-EM structure of the yeast B complex at average resolution of 3.9 angstrom | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 23 kDa U4/U6.U5 small nuclear ribonucleoprotein component, 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, ... | | 著者 | Bai, R, Wan, R, Yan, C, Shi, Y. | | 登録日 | 2018-05-16 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structures of the fully assembledSaccharomyces cerevisiaespliceosome before activation
Science, 360, 2018
|
|
6AH0
 
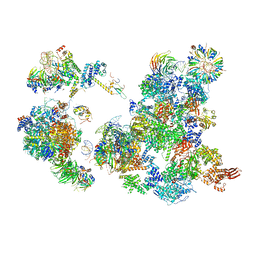 | | The Cryo-EM Structure of the Precusor of Human Pre-catalytic Spliceosome (pre-B complex) | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Zhan, X, Yan, C, Zhang, X, Shi, Y. | | 登録日 | 2018-08-15 | | 公開日 | 2018-11-14 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (5.7 Å) | | 主引用文献 | Structures of the human pre-catalytic spliceosome and its precursor spliceosome.
Cell Res., 28, 2018
|
|
6AHD
 
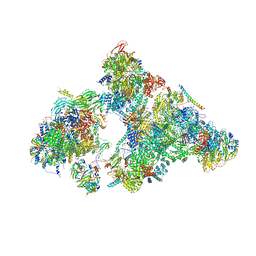 | | The Cryo-EM Structure of Human Pre-catalytic Spliceosome (B complex) at 3.8 angstrom resolution | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, Brr2, U5 small nuclear ribonucleoprotein 200 kDa helicase, ... | | 著者 | Zhan, X, Yan, C, Zhang, X, Shi, Y. | | 登録日 | 2018-08-17 | | 公開日 | 2018-11-14 | | 最終更新日 | 2021-06-30 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structures of the human pre-catalytic spliceosome and its precursor spliceosome.
Cell Res., 28, 2018
|
|
6KZ1
 
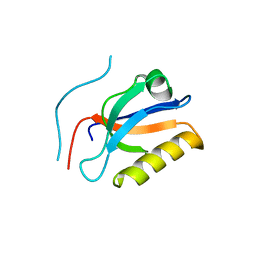 | | Complex structure of Whirlin and Myosin XVa | | 分子名称: | Myosin XVa, Whirlin | | 著者 | Lin, L, Wang, M, Shi, Y, Zhu, J, Zhang, R. | | 登録日 | 2019-09-22 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.694 Å) | | 主引用文献 | Phase separation-mediated condensation of Whirlin-Myo15-Eps8 stereocilia tip complex.
Cell Rep, 34, 2021
|
|
5Z56
 
 | | cryo-EM structure of a human activated spliceosome (mature Bact) at 5.1 angstrom. | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | 著者 | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | 登録日 | 2018-01-17 | | 公開日 | 2018-09-19 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
5Z57
 
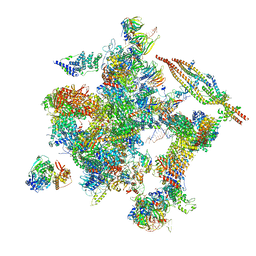 | | Cryo-EM structure of the human activated spliceosome (late Bact) at 6.5 angstrom | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ALANINE, BUD13 homolog, ... | | 著者 | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | 登録日 | 2018-01-17 | | 公開日 | 2018-09-19 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
