7FFH
 
 | | Diarylpentanoid-producing polyketide synthase (N199L mutant) | | Descriptor: | Type III polyketide synthase | | Authors: | Morita, H, Wong, C.P, Liu, Q, Takeshi, K, Lee, Y, Nakashima, Y. | | Deposit date: | 2021-07-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of a diarylpentanoid-producing polyketide synthase revealing an unusual biosynthetic pathway of 2-(2-phenylethyl)chromones in agarwood.
Nat Commun, 13, 2022
|
|
3VN0
 
 | |
8H87
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR2 in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR2, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
8H86
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR1, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
7DDR
 
 | | Ancestral myoglobin aMbSp of Puijila Darwini relative (imidazol ligand) | | Descriptor: | Ancestral myoglobin aMbSp, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Isogai, Y, Imamura, H, Nakae, S, Sumi, T, Takahashi, K, Shirai, T. | | Deposit date: | 2020-10-29 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Common and unique strategies of myoglobin evolution for deep-sea adaptation of diving mammals.
Iscience, 24, 2021
|
|
7DDS
 
 | | Ancestral myoglobin aMbSp of Puijila Darwini relative | | Descriptor: | Ancestral myoglobin aMbSp, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Isogai, Y, Imamura, H, Nakae, S, Sumi, T, Takahashi, K, Shirai, T. | | Deposit date: | 2020-10-29 | | Release date: | 2021-09-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Common and unique strategies of myoglobin evolution for deep-sea adaptation of diving mammals.
Iscience, 24, 2021
|
|
8IU0
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 H225F mutant in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR1, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Nakamura, S, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2023-03-23 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
1IS1
 
 | | Crystal structure of ribosome recycling factor from Vibrio parahaemolyticus | | Descriptor: | RIBOSOME RECYCLING FACTOR | | Authors: | Nakano, H, Yamaichi, Y, Uchiyama, S, Yoshida, T, Nishina, K, Kato, H, Ohkubo, T, Honda, T, Yamagata, Y, Kobayashi, Y. | | Deposit date: | 2001-11-05 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and binding mode of a ribosome recycling factor (RRF) from mesophilic bacterium
J.BIOL.CHEM., 278, 2003
|
|
1IQA
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF MOUSE RANK LIGAND | | Descriptor: | RECEPTOR ACTIVATOR OF NUCLEAR FACTOR KAPPA B LIGAND | | Authors: | Ito, S, Wakabayashi, K, Ubukata, O, Hayashi, S, Okada, F, Hata, T. | | Deposit date: | 2001-07-11 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the extracellular domain of mouse RANK ligand at 2.2-A resolution.
J.Biol.Chem., 277, 2002
|
|
8H25
 
 | | Lacticaseibacillus casei GH35 beta-galactosidase LBCZ_0230 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-galactosidase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Saburi, W, Ota, T, Kato, K, Tagami, T, Yamashita, K, Yao, M, Mori, H. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Function and Structure of Lacticaseibacillus casei GH35 beta-Galactosidase LBCZ_0230 with High Hydrolytic Activity to Lacto- N -biose I and Galacto- N -biose.
J Appl Glycosci (1999), 70, 2023
|
|
1B7R
 
 | | VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Ota, M, Ogasahara, K, Yamagata, Y, Nishikawa, K, Yutani, K. | | Deposit date: | 1999-01-25 | | Release date: | 1999-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental verification of the 'stability profile of mutant protein' (SPMP) data using mutant human lysozymes.
Protein Eng., 12, 1999
|
|
6JU6
 
 | | Aspergillus oryzae active-tyrosinase copper-depleted C92A mutant | | Descriptor: | NITRATE ION, Tyrosinase | | Authors: | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-04-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JUD
 
 | | Radiation damage in Aspergillus oryzae pro-tyrosinase oxygen-bound C92A/H103F mutant | | Descriptor: | COPPER (II) ION, PEROXIDE ION, Tyrosinase | | Authors: | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-04-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8HLB
 
 | | Cryo-EM structure of biparatopic antibody Bp109-92 in complex with TNFR2 | | Descriptor: | TR109 heavy chain, TR109 light chain, TR92 heavy chain, ... | | Authors: | Akiba, H, Fujita, J, Ise, T, Nishiyama, K, Miyata, T, Kato, T, Namba, K, Ohno, H, Kamada, H, Nagata, S, Tsumoto, K. | | Deposit date: | 2022-11-29 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Development of a 1:1-binding biparatopic anti-TNFR2 antagonist by reducing signaling activity through epitope selection.
Commun Biol, 6, 2023
|
|
6K7H
 
 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1 state class2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Cell cycle control protein 50A, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-06-07 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
6JUC
 
 | | Aspergillus oryzae pro-tyrosinase oxygen-bound C92A/H103F mutant | | Descriptor: | COPPER (II) ION, PEROXIDE ION, Tyrosinase | | Authors: | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-04-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JU5
 
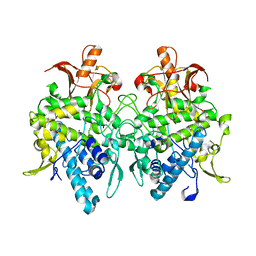 | | Aspergillus oryzae pro-tyrosinase C92A/F513Y mutant | | Descriptor: | COPPER (II) ION, Tyrosinase | | Authors: | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-04-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6K7I
 
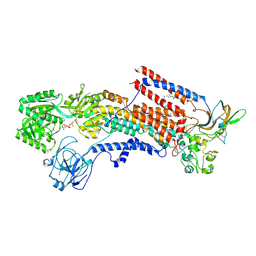 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1-ATP state class2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Cell cycle control protein 50A, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-06-07 | | Release date: | 2019-08-28 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
6K7N
 
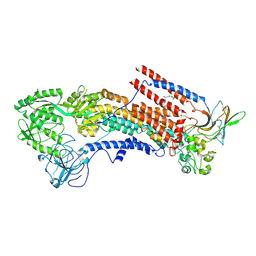 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1P state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Cell cycle control protein 50A, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-06-07 | | Release date: | 2019-08-28 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
1B7P
 
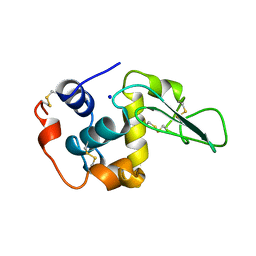 | | VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Ota, M, Ogasahara, K, Yamagata, Y, Nishikawa, K, Yutani, K. | | Deposit date: | 1998-05-08 | | Release date: | 1999-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Experimental verification of the 'stability profile of mutant protein' (SPMP) data using mutant human lysozymes.
Protein Eng., 12, 1999
|
|
8W7N
 
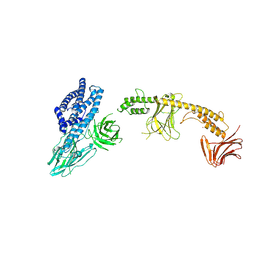 | | Crystal structure of the in-cell Cry1Aa purified from Bacillus thuringiensis | | Descriptor: | Pesticidal crystal protein Cry1Aa, UNKNOWN ATOM OR ION | | Authors: | Tanaka, J, Abe, S, Hayakawa, T, Kojima, M, Yamashita, K, Hirata, K, Ueno, T. | | Deposit date: | 2023-08-31 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of the in-cell Cry1Aa purified from Bacillus thuringiensis.
Biochem.Biophys.Res.Commun., 685, 2023
|
|
1B7S
 
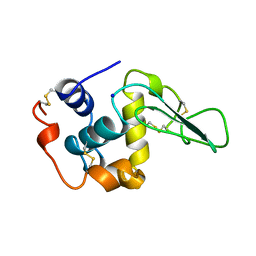 | | VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Ota, M, Ogasahara, K, Yamagata, Y, Nishikawa, K, Yutani, K. | | Deposit date: | 1999-01-25 | | Release date: | 1999-02-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Experimental verification of the 'stability profile of mutant protein' (SPMP) data using mutant human lysozymes.
Protein Eng., 12, 1999
|
|
5VM9
 
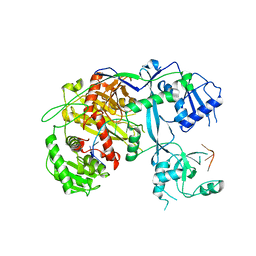 | | Human Argonaute3 bound to guide RNA | | Descriptor: | Protein argonaute-3, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*UP*U)-3'), RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*UP*U)-3') | | Authors: | Park, M.S, Nakanishi, K. | | Deposit date: | 2017-04-26 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Human Argonaute3 has slicer activity.
Nucleic Acids Res., 45, 2017
|
|
7E2E
 
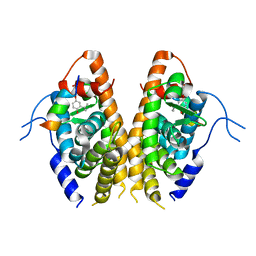 | | Crystal structure of the Estrogen-Related Receptor alpha (ERRalpha) ligand-binding domain (LBD) in complex with an agonist DS45500853 and a PGC-1alpha peptide | | Descriptor: | 1-[4-(3-tert-butyl-4-oxidanyl-phenoxy)phenyl]ethanone, IODIDE ION, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, ... | | Authors: | Ito, S, Shinozuka, T, Kimura, T, Izumi, M, Wakabayashi, K. | | Deposit date: | 2021-02-05 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of a Novel Class of ERR alpha Agonists.
Acs Med.Chem.Lett., 12, 2021
|
|
6JU7
 
 | | Aspergillus oryzae active-tyrosinase copper-depleted C92A mutant complexed with L-tyrosine | | Descriptor: | NITRATE ION, TYROSINE, Tyrosinase | | Authors: | Fujieda, N, Umakoshi, K, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-04-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Copper-Oxygen Dynamics in the Tyrosinase Mechanism.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
