1K1V
 
 | | Solution Structure of the DNA-Binding Domain of MafG | | Descriptor: | MafG | | Authors: | Kusunoki, H, Motohashi, H, Katsuoka, F, Morohashi, A, Yamamoto, M, Tanaka, T. | | Deposit date: | 2001-09-25 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of MafG.
Nat.Struct.Biol., 9, 2002
|
|
3VJH
 
 | | Human PPAR GAMMA ligand binding domain in complex with JKPL35 | | Descriptor: | (2S)-2-[4-methoxy-3-({[4-(trifluoromethyl)benzoyl]amino}methyl)benzyl]pentanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Tomioka, D, Kuwabara, N, Hashimoto, H, Sato, M, Shimizu, T. | | Deposit date: | 2011-10-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Peroxisome proliferator-activated receptors (PPARs) have multiple binding points that accommodate ligands in various conformations: phenylpropanoic acid-type PPAR ligands bind to PPAR in different conformations, depending on the subtype.
J.Med.Chem., 55, 2012
|
|
6JP6
 
 | | The X-ray structure of yeast tRNA methyltransferase complex of Trm7 and Trm734 essential for 2'-O-methylation at the first position of anticodon in specific tRNAs | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, tRNA (cytidine(34)/guanosine(34)-2'-O)-methyltransferase, ... | | Authors: | Hirata, A, Okada, K, Yoshii, K, Shiraisi, H, Saijo, S, Yonezawa, K, Sihimzu, N, Hori, H. | | Deposit date: | 2019-03-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Structure of tRNA methyltransferase complex of Trm7 and Trm734 reveals a novel binding interface for tRNA recognition.
Nucleic Acids Res., 47, 2019
|
|
4GTW
 
 | | Crystal structure of mouse Enpp1 in complex with AMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Kato, K, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-29 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Enpp1, an extracellular glycoprotein involved in bone mineralization and insulin signaling.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GTZ
 
 | | Crystal structure of mouse Enpp1 in complex with CMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kato, K, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-29 | | Release date: | 2012-11-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.191 Å) | | Cite: | Crystal structure of Enpp1, an extracellular glycoprotein involved in bone mineralization and insulin signaling.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7DEN
 
 | | Crystal structure of P.aeruginosa LpxC in complex with inhibitor | | Descriptor: | 4-[(1~{R},5~{S})-6-[2-[4-[3-[[2-[(1~{S})-1-oxidanylethyl]imidazol-1-yl]methyl]-1,2-oxazol-5-yl]phenyl]ethynyl]-3-azabicyclo[3.1.0]hexan-3-yl]butanoic acid, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Mima, M, Ushiyama, F, Takashima, H. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Lead optimization of 2-hydroxymethyl imidazoles as non-hydroxamate LpxC inhibitors: Discovery of TP0586532.
Bioorg.Med.Chem., 30, 2020
|
|
4GTY
 
 | | Crystal structure of mouse Enpp1 in complex with GMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kato, K, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-29 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal structure of Enpp1, an extracellular glycoprotein involved in bone mineralization and insulin signaling.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GTX
 
 | | Crystal structure of mouse Enpp1 in complex with TMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kato, K, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2012-08-29 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Crystal structure of Enpp1, an extracellular glycoprotein involved in bone mineralization and insulin signaling.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VJI
 
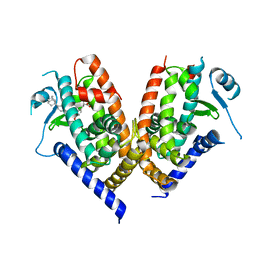 | | Human PPAR gamma ligand binding domain in complex with JKPL53 | | Descriptor: | (2S)-2-{4-butoxy-3-[({4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]benzoyl}amino)methyl]benzyl}butanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Tomioka, D, Kuwabara, N, Hashimoto, H, Sato, M, Shimizu, T. | | Deposit date: | 2011-10-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Peroxisome proliferator-activated receptors (PPARs) have multiple binding points that accommodate ligands in various conformations: phenylpropanoic acid-type PPAR ligands bind to PPAR in different conformations, depending on the subtype.
J.Med.Chem., 55, 2012
|
|
6L9C
 
 | | Neutron structure of copper amine oxidase from Arthrobacter glibiformis at pD 7.4 | | Descriptor: | COPPER (II) ION, Phenylethylamine oxidase, SODIUM ION | | Authors: | Murakawa, T, Kurihara, K, Shoji, M, Shibazaki, C, Sunami, T, Tamada, T, Yano, N, Yamada, T, Kusaka, K, Suzuki, M, Shigeta, Y, Kuroki, R, Hayashi, H, Yano, Y, Tanizawa, K, Adachi, M, Okajima, T. | | Deposit date: | 2019-11-08 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | NEUTRON DIFFRACTION (1.14 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallography of copper amine oxidase reveals keto/enolate interconversion of the quinone cofactor and unusual proton sharing.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3PIO
 
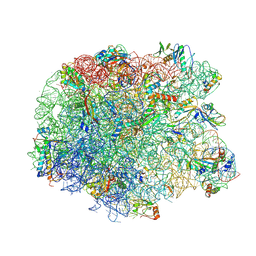 | | Crystal structure of the synergistic antibiotic pair lankamycin and lankacidin in complex with the large ribosomal subunit | | Descriptor: | 50S ribosomal protein L11, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Belousoff, M.J, Shapira, T, Bashan, A, Zimmerman, E, Arakawa, K, Kinashi, H, Rozenberg, H, Yonath, A. | | Deposit date: | 2010-11-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2473 Å) | | Cite: | Crystal structure of the synergistic antibiotic pair, lankamycin and lankacidin, in complex with the large ribosomal subunit.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PIP
 
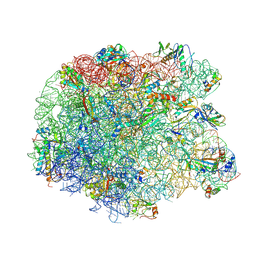 | | Crystal structure of the synergistic antibiotic pair lankamycin and lankacidin in complex with the large ribosomal subunit | | Descriptor: | 50S ribosomal protein L11, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Belousoff, M.J, Shapira, T, Bashan, A, Zimmerman, E, Kinashi, H, Rozenberg, H, Yonath, A. | | Deposit date: | 2010-11-07 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Crystal structure of the synergistic antibiotic pair, lankamycin and lankacidin, in complex with the large ribosomal subunit.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7EFQ
 
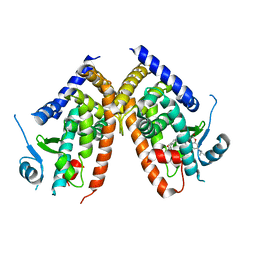 | | Crystal structure of hPPARgamma ligand binding domain complexed with rosiglitazone-based fluorescence probe | | Descriptor: | (5S)-5-[[4-[2-[[7-(diethylamino)-2-oxidanylidene-chromen-4-yl]-methyl-amino]ethoxy]phenyl]methyl]-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma | | Authors: | Yoshikawa, C, Ishida, H, Ohashi, N, Itoh, T. | | Deposit date: | 2021-03-23 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis of a Coumarin-Based PPAR gamma Fluorescence Probe for Competitive Binding Assay.
Int J Mol Sci, 22, 2021
|
|
8W8M
 
 | | Cryo-EM structure of helical filament of MyD88 TIR | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Kasai, K, Imamura, K, Narita, A, Makino, F, Miyata, T, Kato, T, Namba, K, Onishi, H, Tochio, H. | | Deposit date: | 2023-09-04 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Signaling adopter protein in a self-assembled form
To Be Published
|
|
7FI3
 
 | | Archaeal oligopeptide permease A (OppA) from Thermococcus kodakaraensis in complex with an endogenous pentapeptide | | Descriptor: | ABC-type dipeptide/oligopeptide transport system, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Yokoyama, H, Kamei, N, Konishi, K, Hara, K, Hashimoto, H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for peptide recognition by archaeal oligopeptide permease A.
Proteins, 90, 2022
|
|
1G7X
 
 | | ASPARTATE AMINOTRANSFERASE ACTIVE SITE MUTANT N194A/R292L/R386L | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Mizuguchi, H, Hayashi, H, Okada, K, Miyahara, I, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2000-11-15 | | Release date: | 2000-11-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Strain is more important than electrostatic interaction in controlling the pKa of the catalytic group in aspartate aminotransferase.
Biochemistry, 40, 2001
|
|
4KRU
 
 | |
7CP6
 
 | | Crystal structure of FqzB | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, IODIDE ION, MAK1-like monooxygenase, ... | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Kishimoto, S, Watanabe, K. | | Deposit date: | 2020-08-06 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analyses of a Spiro-Carbon-Forming, Highly Promiscuous Epoxidase from Fungal Natural Product Biosynthesis.
Biochemistry, 59, 2020
|
|
1G4V
 
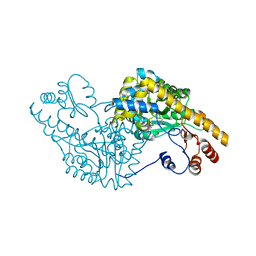 | | ASPARTATE AMINOTRANSFERASE ACTIVE SITE MUTANT N194A/Y225F | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Mizuguchi, H, Hayashi, H, Okada, K, Miyahara, I, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2000-10-28 | | Release date: | 2000-11-22 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Strain is more important than electrostatic interaction in controlling the pKa of the catalytic group in aspartate aminotransferase.
Biochemistry, 40, 2001
|
|
1G7W
 
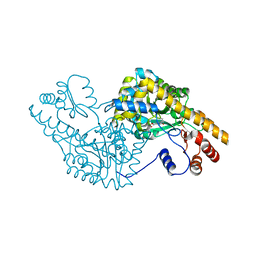 | | ASPARTATE AMINOTRANSFERASE ACTIVE SITE MUTANT N194A/R386L | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Mizuguchi, H, Hayashi, H, Okada, K, Miyahara, I, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2000-11-15 | | Release date: | 2000-11-29 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Strain is more important than electrostatic interaction in controlling the pKa of the catalytic group in aspartate aminotransferase.
Biochemistry, 40, 2001
|
|
1GDV
 
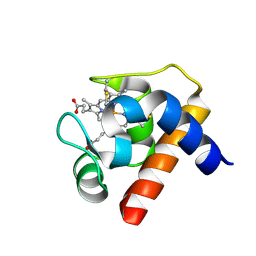 | | CRYSTAL STRUCTURE OF CYTOCHROME C6 FROM RED ALGA PORPHYRA YEZOENSIS AT 1.57 A RESOLUTION | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Yamada, S, Park, S.-Y, Shimizu, H, Shiro, Y, Oku, T. | | Deposit date: | 2000-10-06 | | Release date: | 2001-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure of cytochrome c6 from the red alga Porphyra yezoensis at 1. 57 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1G0C
 
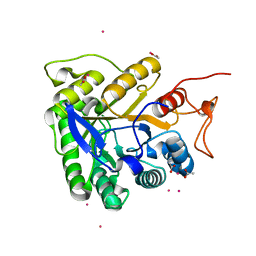 | | ALKALINE CELLULASE K CATALYTIC DOMAIN-CELLOBIOSE COMPLEX | | Descriptor: | ACETIC ACID, CADMIUM ION, ENDOGLUCANASE, ... | | Authors: | Shirai, T, Ishida, H, Noda, J, Yamane, T, Ozaki, K, Hakamada, Y, Ito, S. | | Deposit date: | 2000-10-05 | | Release date: | 2001-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of alkaline cellulase K: insight into the alkaline adaptation of an industrial enzyme.
J.Mol.Biol., 310, 2001
|
|
7CP7
 
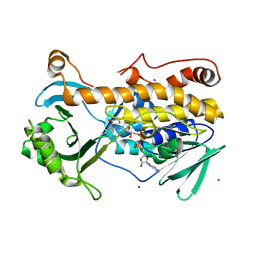 | | Crystal structure of FqzB, native proteins | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, IODIDE ION, MAK1-like monooxygenase | | Authors: | Hara, K, Hashimoto, H, Matsushita, T, Kishimoto, S, Watanabe, K. | | Deposit date: | 2020-08-06 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Analyses of a Spiro-Carbon-Forming, Highly Promiscuous Epoxidase from Fungal Natural Product Biosynthesis.
Biochemistry, 59, 2020
|
|
1G01
 
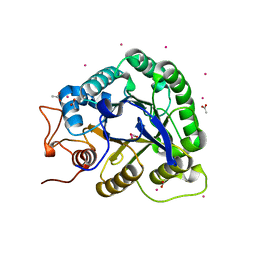 | | ALKALINE CELLULASE K CATALYTIC DOMAIN | | Descriptor: | ACETIC ACID, CADMIUM ION, ENDOGLUCANASE | | Authors: | Shirai, T, Ishida, H, Noda, J, Yamane, T, Ozaki, K, Hakamada, Y, Ito, S. | | Deposit date: | 2000-10-05 | | Release date: | 2001-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of alkaline cellulase K: insight into the alkaline adaptation of an industrial enzyme.
J.Mol.Biol., 310, 2001
|
|
4KRT
 
 | |
