7YTW
 
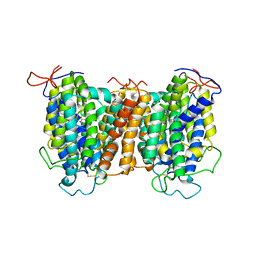 | | Structural basis of vitamin C recognition and transport by mammalian SVCT1 transporter | | Descriptor: | ASCORBIC ACID, SODIUM ION, Solute carrier family 23 member 1 | | Authors: | She, J, Wang, M, He, J, Zhang, K, Li, S. | | Deposit date: | 2022-08-16 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of vitamin C recognition and transport by mammalian SVCT1 transporter.
Nat Commun, 14, 2023
|
|
7YTY
 
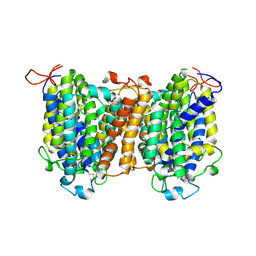 | | Mouse SVCT1 in an apo state | | Descriptor: | Solute carrier family 23 member 1 | | Authors: | She, J, Wang, M, He, J, Zhang, K, Li, S. | | Deposit date: | 2022-08-16 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of vitamin C recognition and transport by mammalian SVCT1 transporter.
Nat Commun, 14, 2023
|
|
6C9A
 
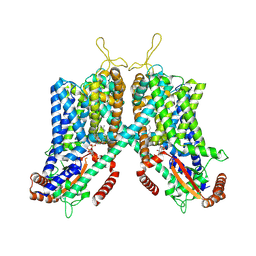 | | Cryo-EM structure of mouse TPC1 channel in the PtdIns(3,5)P2-bound state | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | She, J, Guo, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2018-01-25 | | Release date: | 2018-04-04 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the voltage and phospholipid activation of the mammalian TPC1 channel.
Nature, 556, 2018
|
|
6C96
 
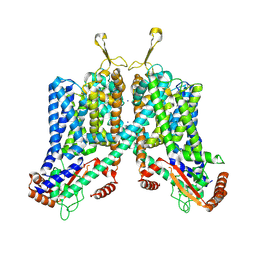 | | Cryo-EM structure of mouse TPC1 channel in the apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | Authors: | She, J, Guo, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2018-01-25 | | Release date: | 2018-04-04 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the voltage and phospholipid activation of the mammalian TPC1 channel.
Nature, 556, 2018
|
|
8IOW
 
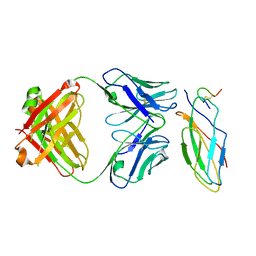 | |
8J6F
 
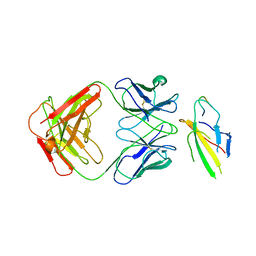 | | Cryo-EM structure of the Tocilizumab Fab/IL-6R complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Tocilizumab Fab, IL6R-D2 peptide, ... | | Authors: | She, J, Chen, L. | | Deposit date: | 2023-04-25 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of tocilizumab and sarilumab Fabs in complex with IL-6R
To Be Published
|
|
6NQ2
 
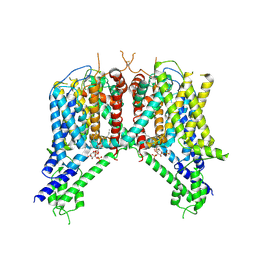 | | Cryo-EM structure of human TPC2 channel in the ligand-bound closed state | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, Two pore calcium channel protein 2 | | Authors: | She, J, Zeng, W, Guo, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2019-01-18 | | Release date: | 2019-03-27 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural mechanisms of phospholipid activation of the human TPC2 channel.
Elife, 8, 2019
|
|
6NQ1
 
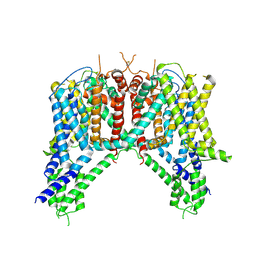 | | Cryo-EM structure of human TPC2 channel in the apo state | | Descriptor: | Two pore calcium channel protein 2 | | Authors: | She, J, Zeng, W, Guo, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2019-01-18 | | Release date: | 2019-03-27 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural mechanisms of phospholipid activation of the human TPC2 channel.
Elife, 8, 2019
|
|
6NQ0
 
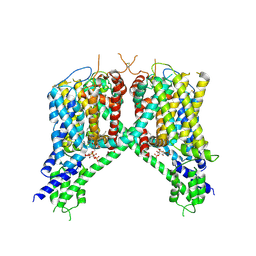 | | Cryo-EM structure of human TPC2 channel in the ligand-bound open state | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, Two pore calcium channel protein 2 | | Authors: | She, J, Zeng, W, Guo, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2019-01-18 | | Release date: | 2019-03-27 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural mechanisms of phospholipid activation of the human TPC2 channel.
Elife, 8, 2019
|
|
4U34
 
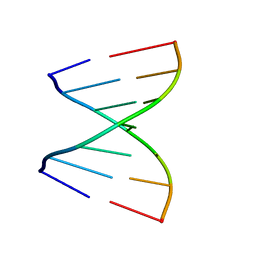 | | Crystal Structures of RNA Duplexes Containing 2-thio-Uridine | | Descriptor: | RNA (5'-R(*GP*GP*AP*GP*CP*UP*A)-3'), RNA (5'-R(*UP*AP*GP*CP*(SUR)P*CP*C-3') | | Authors: | Sheng, J, Larsen, A, Heuberger, B, Blain, J.C, Szostak, J.W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure Studies of RNA Duplexes Containing s(2)U:A and s(2)U:U Base Pairs.
J.Am.Chem.Soc., 136, 2014
|
|
3LTR
 
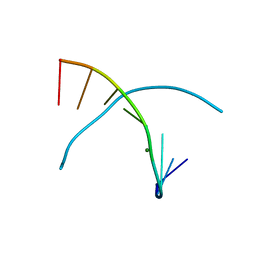 | | 5-OMe-dU containing DNA 8mer | | Descriptor: | 5'-D(*GP*(UMS)P*GP*(T5O)P*AP*CP*AP*C)-3', MAGNESIUM ION | | Authors: | Sheng, J, Hassan, A.E.A, Zhang, W, Gan, J, Huang, Z. | | Deposit date: | 2010-02-16 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Hydrogen bond formation between the naturally modified nucleobase and phosphate backbone.
Nucleic Acids Res., 40, 2012
|
|
4U38
 
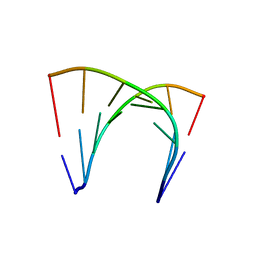 | | RNA duplex containing UU mispair | | Descriptor: | RNA (5'-R(*GP*GP*UP*GP*CP*UP*A)-3'), RNA (5'-R(*UP*AP*GP*CP*UP*CP*C)-3') | | Authors: | Sheng, J, Larsen, A, Heuberger, B, Blain, J.C, Szostak, J.W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure Studies of RNA Duplexes Containing s(2)U:A and s(2)U:U Base Pairs.
J.Am.Chem.Soc., 136, 2014
|
|
4U35
 
 | | Crystal Structures of RNA Duplexes Containing 2-thio-Uridine | | Descriptor: | RNA (5'-R(*GP*GP*UP*GP*CP*UP*A)-3'), RNA (5'-R(*UP*AP*GP*CP*(SUR)P*CP*C-3') | | Authors: | Sheng, J, Larsen, A, Heuberger, B, Blain, J.C, Szostak, J.W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure Studies of RNA Duplexes Containing s(2)U:A and s(2)U:U Base Pairs.
J.Am.Chem.Soc., 136, 2014
|
|
4U37
 
 | | Native 7mer-RNA duplex | | Descriptor: | RNA (5'-R(*GP*GP*AP*GP*CP*UP*A)-3'), RNA (5'-R(*UP*AP*GP*CP*UP*CP*C)-3') | | Authors: | Sheng, J, Larsen, A, Heuberger, B, Blain, J.C, Szostak, J.W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure Studies of RNA Duplexes Containing s(2)U:A and s(2)U:U Base Pairs.
J.Am.Chem.Soc., 136, 2014
|
|
3LTU
 
 | | 5-SeMe-dU containing DNA 8mer | | Descriptor: | 5'-D(*GP*(UMS)P*GP*(T5S)P*AP*CP*AP*C)-3' | | Authors: | Sheng, J, Hassan, A.E.A, Zhang, W, Gan, J, Huang, Z. | | Deposit date: | 2010-02-16 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Synthesis of Pyrimidine Modified Seleno-DNA as a Novel Approach to Antisense Candidate
Chemistryselect, 8, 2023
|
|
8G3G
 
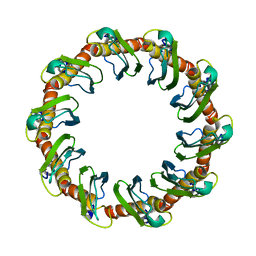 | | CryoEM structure of yeast recombination mediator Rad52 | | Descriptor: | DNA repair and recombination protein RAD52 | | Authors: | Deveryshetty, J, Basore, K, Rau, M, Fitzpatrick, J.A.J, Antony, E. | | Deposit date: | 2023-02-07 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Yeast Rad52 is a homodecamer and possesses BRCA2-like bipartite Rad51 binding modes.
Nat Commun, 14, 2023
|
|
8HZ5
 
 | |
8HZ4
 
 | |
5JJD
 
 | |
6WF2
 
 | | Crystal structure of mouse SCD1 with a diiron center | | Descriptor: | Acyl-CoA desaturase 1, FE (III) ION, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name) | | Authors: | Shen, J, Zhou, M. | | Deposit date: | 2020-04-03 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structure and Mechanism of a Unique Diiron Center in Mammalian Stearoyl-CoA Desaturase.
J.Mol.Biol., 432, 2020
|
|
7S8U
 
 | |
8DL7
 
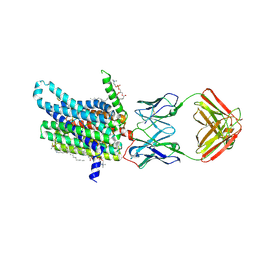 | | Cryo-EM structure of human ferroportin/slc40 bound to minihepcidin PR73 in nanodisc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 11F9 heavy-chain, 11F9 light-chain, ... | | Authors: | Shen, J, Wilbon, A.S, Pan, Y, Zhou, M. | | Deposit date: | 2022-07-07 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of ferroportin inhibition by minihepcidin PR73.
Plos Biol., 21, 2023
|
|
8DL8
 
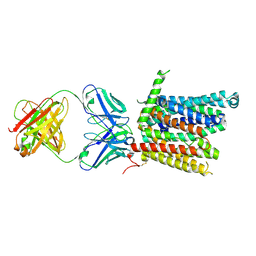 | | Cryo-EM structure of human ferroportin/slc40 bound to Co2+ in nanodisc | | Descriptor: | 11F9 heavy-chain, 11F9 light-chain, COBALT (II) ION, ... | | Authors: | Shen, J, Wilbon, A.S, Pan, Y, Zhou, M. | | Deposit date: | 2022-07-07 | | Release date: | 2022-12-07 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of ferroportin inhibition by minihepcidin PR73.
Plos Biol., 21, 2023
|
|
8DL6
 
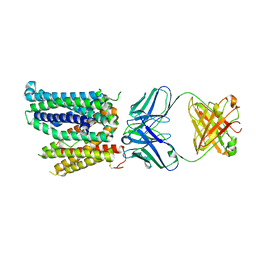 | | Cryo-EM structure of human ferroportin/slc40 bound to Ca2+ in nanodisc | | Descriptor: | 11F9 heavy-chain, 11F9 light-chain, CALCIUM ION, ... | | Authors: | Shen, J, Wilbon, A.S, Pan, Y, Zhou, M. | | Deposit date: | 2022-07-07 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of Ca 2+ transport by ferroportin.
Elife, 12, 2023
|
|
6VYH
 
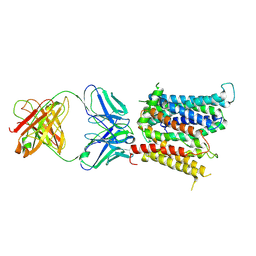 | | Cryo-EM structure of SLC40/ferroportin in complex with Fab | | Descriptor: | 11F9 heavy-chain, 11F9 light-chain, COBALT (II) ION, ... | | Authors: | Shen, J, Ren, Z, Pan, Y, Gao, S, Yan, N, Zhou, M. | | Deposit date: | 2020-02-26 | | Release date: | 2020-11-11 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of ion transport and inhibition in ferroportin.
Nat Commun, 11, 2020
|
|
