3TTR
 
 | | Crystal structure of C-lobe of bovine lactoferrin complexed with Lidocaine at 2.27 A resolution | | Descriptor: | 2-(diethylamino)-N-(2,6-dimethylphenyl)ethanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamini, S, Gautam, L, Singh, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-09-15 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of C-lobe of bovine lactoferrin complexed with Lidocaine at 2.27 A resolution
To be Published
|
|
3TUS
 
 | | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with Meta-hydroxy benzoic acid at 2.5 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-HYDROXYBENZOIC ACID, ... | | Authors: | Shukla, P.K, Gautam, L, Singh, A, Kaushik, S, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-09-18 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with Meta-hydroxy benzoic acid at 2.5 A Resolution
To be Published
|
|
5ILX
 
 | | Crystal structure of Ribosome inactivating protein from Momordica balsamina with Uracil at 1.70 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome inactivating protein, ... | | Authors: | Singh, P.K, Singh, A, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-03-05 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Ribosome inactivating protein from Momordica balsamina with Uracil at 1.70 Angstrom resolution
To Be Published
|
|
8I11
 
 | |
1OYO
 
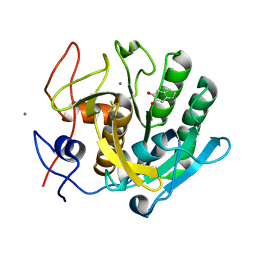 | | Regulation of protease activity by melanin: Crystal structure of the complex formed between proteinase K and melanin monomers at 2.0 resolution | | Descriptor: | 3H-INDOLE-5,6-DIOL, CALCIUM ION, Proteinase K | | Authors: | Singh, N, Sharma, S, Kumar, S, Raman, G, Singh, T.P. | | Deposit date: | 2003-04-06 | | Release date: | 2003-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Regulation of protease activity by melanin: Crystal structure of the complex formed between proteinase K and melanin monomers at 2.0 resolution
To be Published
|
|
5HBC
 
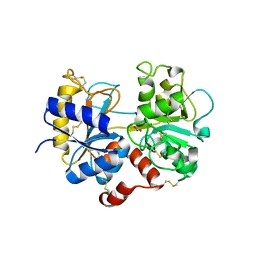 | | Intermediate structure of iron-saturated C-lobe of bovine lactoferrin at 2.79 Angstrom resolution indicates the softening of iron coordination | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, FE (III) ION, ... | | Authors: | Singh, A, Rastogi, N, Singh, P.K, Tyagi, T.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-12-31 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure of iron saturated C-lobe of bovine lactoferrin at pH 6.8 indicates a weakening of iron coordination
Proteins, 84, 2016
|
|
8IL9
 
 | |
1Q7A
 
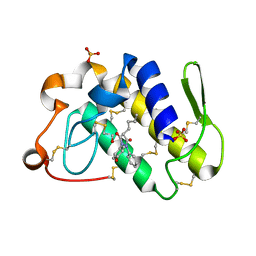 | | Crystal structure of the complex formed between russell's viper phospholipase A2 and an antiinflammatory agent oxyphenbutazone at 1.6A resolution | | Descriptor: | 4-BUTYL-1-(4-HYDROXYPHENYL)-2-PHENYLPYRAZOLIDINE-3,5-DIONE, METHANOL, Phospholipase A2 VRV-PL-VIIIa, ... | | Authors: | Singh, N, Jabeen, T, Sharma, S, Singh, T.P. | | Deposit date: | 2003-08-17 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Phospholipase A2 as a target protein for nonsteroidal anti-inflammatory drugs (NSAIDS): crystal structure of the complex formed between phospholipase A2 and oxyphenbutazone at 1.6 A resolution.
Biochemistry, 43, 2004
|
|
8IYN
 
 | |
8J68
 
 | |
8EYO
 
 | | Crystal Structure of Human Mitochondrial NADP+ Malic Enzyme 3 with NADP bound | | Descriptor: | CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent malic enzyme, ... | | Authors: | Shaffer, P.L, Grell, T.A.J, Mason, M, Thompson, A.A, Riley, D, Wagner, M.V, Steele, R, Ortiz-Meoz, R, Wadia, J, Sharma, S, Yu, X. | | Deposit date: | 2022-10-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Integrative structural and functional analysis of human malic enzyme 3: A potential therapeutic target for pancreatic cancer.
Heliyon, 8, 2022
|
|
8EYN
 
 | | Crystal Structure of Human Mitochondrial NADP+ Malic Enzyme 3 in Apo Form | | Descriptor: | CITRIC ACID, NADP-dependent malic enzyme, mitochondrial | | Authors: | Shaffer, P.L, Grell, T.A.J, Mason, M, Thompson, A.A, Riley, D, Wagner, M.V, Steele, R, Ortiz-Meoz, R, Wadia, J, Sharma, S, Yu, X. | | Deposit date: | 2022-10-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Integrative structural and functional analysis of human malic enzyme 3: A potential therapeutic target for pancreatic cancer.
Heliyon, 8, 2022
|
|
5ILW
 
 | | Crystal structure of the complex of type 1 Ribosome inactivating protein from Momordica balsamina with Uridine at 1.97 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Singh, P.K, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-03-05 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein from Momordica balsamina with uracil and uridine.
Proteins, 87, 2019
|
|
3TGY
 
 | | Crystal structure of the complex of Bovine Lactoperoxidase with Ascorbic acid at 2.35 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamini, S, Singh, R.P, Singh, A.K, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-08-18 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of bovine lactoperoxidase with a partially linked heme moiety at 1.98 angstrom resolution.
Biochim.Biophys.Acta, 1865, 2017
|
|
5Y48
 
 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with Pyrimidine-2,4-dione at 1.70 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome inactivating protein, URACIL | | Authors: | Singh, P.K, Pandey, S, Iqbal, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-08-01 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein from Momordica balsamina with uracil and uridine.
Proteins, 87, 2019
|
|
2DSV
 
 | | Interactions of protective signalling factor with chitin-like polysaccharide: Crystal structure of the complex between signalling protein from sheep (SPS-40) and a hexasaccharide at 2.5A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, alpha-D-mannopyranose-(1-4)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Srivastava, D.B, Ethayathulla, A.S, Kumar, J, Singh, N, Sharma, S, Bhushan, A, Srinivasan, A, Singh, T.P. | | Deposit date: | 2006-07-07 | | Release date: | 2006-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Carbohydrate binding properties and carbohydrate induced conformational switch in sheep secretory glycoprotein (SPS-40): crystal structures of four complexes of SPS-40 with chitin-like oligosaccharides
J.Struct.Biol., 158, 2007
|
|
2DO2
 
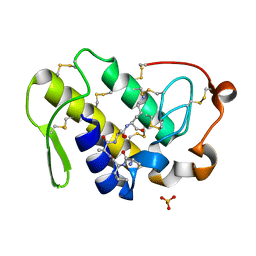 | | Design of specific inhibitors of phospholipase A2: Crystal structure of the complex formed between a group II Cys 49 phospholipase A2 and a designed pentapeptide Ala-Leu-Ala-Ser-Lys at 2.6A resolution | | Descriptor: | Ala-Leu-Ala-Ser-Lys, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Singh, N, Sharma, S, Somvanshi, R.K, Dey, S, Singh, T.P. | | Deposit date: | 2006-04-27 | | Release date: | 2006-05-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design of specific inhibitors of phospholipase A2: Crystal structure of the complex formed between a group II Cys 49 phospholipase A2 and a designed pentapeptide Ala-Leu-Ala-Ser-Lys at 2.6A resolution
To be Published
|
|
2DQK
 
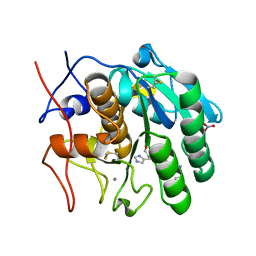 | | Crystal structure of the complex of proteinase K with a specific lactoferrin peptide Val-Leu-Leu-His at 1.93 A resolution | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K, ... | | Authors: | Singh, A.K, Singh, N, Sharma, S, Dey, S, Bhushan, A, Singh, T.P. | | Deposit date: | 2006-05-29 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of the complex of proteinase K with a specific lactoferrin peptide Val-Leu-Leu-His at 1.93 resolution
To be Published
|
|
2DUJ
 
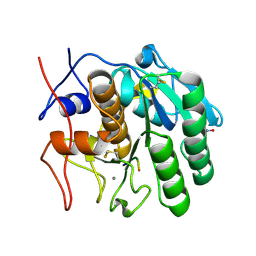 | | Crystal structure of the complex formed between proteinase K and a synthetic peptide Leu-Leu-Phe-Asn-Asp at 1.67 A resolution | | Descriptor: | CALCIUM ION, LLFND, NITRATE ION, ... | | Authors: | Singh, A.K, Singh, N, Somvanshi, R.K, Gupta, D, Sharma, S, Singh, T.P. | | Deposit date: | 2006-07-23 | | Release date: | 2006-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of the complex of proteinase K with a specific lactoferrin peptide Val-Leu-Leu-His at 1.93 A resolution
To be Published
|
|
2DSU
 
 | | Binding of chitin-like polysaccharide to protective signalling factor: Crystal structure of the complex formed between signalling protein from sheep (SPS-40) with a tetrasaccharide at 2.2 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase-3-like protein 1, alpha-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Srivastava, D.B, Ethayathulla, A.S, Kumar, J, Singh, N, Sharma, S, Kaur, P, Bhushan, A, Singh, T.P. | | Deposit date: | 2006-07-07 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Carbohydrate binding properties and carbohydrate induced conformational switch in sheep secretory glycoprotein (SPS-40): crystal structures of four complexes of SPS-40 with chitin-like oligosaccharides
J.Struct.Biol., 158, 2007
|
|
3PTL
 
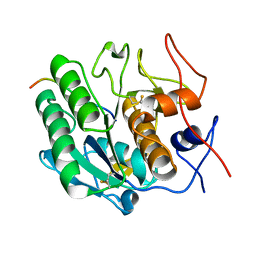 | | Crystal structure of proteinase K inhibited by a lactoferrin nonapeptide, Lys-Gly-Glu-Ala-Asp-Ala-Leu-Ser-Leu-Asp at 1.3 A resolution. | | Descriptor: | 10-mer peptide from Lactoferrin, Proteinase K | | Authors: | Shukla, P.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-12-03 | | Release date: | 2010-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of proteinase K inhibited by a lactoferrin nonapeptide, Lys-Gly-Glu-Ala-Asp-Ala-Leu-Ser-Leu-Asp at 1.3 A resolution.
TO BE PUBLISHED
|
|
3RVG
 
 | | Crystals structure of Jak2 with a 1-amino-5H-pyrido[4,3-b]indol-4-carboxamide inhibitor | | Descriptor: | 1-(cyclohexylamino)-7-(1-methyl-1H-pyrazol-4-yl)-5H-pyrido[4,3-b]indole-4-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Lim, J, Taoka, B, Otte, R.D, Spencer, K, Dinsmore, C.J, Altman, M.D, Chan, G, Rosenstein, C, Sharma, S, Su, H.P, Szewczak, A.A, Xu, L, Yin, H, Zugay-Murphy, J, Marshall, C.G, Young, J.R. | | Deposit date: | 2011-05-06 | | Release date: | 2012-03-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Discovery of 1-amino-5H-pyrido[4,3-b]indol-4-carboxamide inhibitors of Janus kinase 2 (JAK2) for the treatment of myeloproliferative disorders.
J.Med.Chem., 54, 2011
|
|
3SJ6
 
 | | Crystal Structure of the complex of type I ribosome inactivating protein from momordica balsamina with 5-(hydroxymethyl)oxalane-2,3,4-triol at 1.6 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Pandey, N, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of a type-1 ribosome inactivating protein from Momordica balsamina in the bound and unbound states.
Biochim.Biophys.Acta, 1824, 2012
|
|
3RKE
 
 | | Crystal Structure of goat Lactoperoxidase complexed with a tightly bound inhibitor, 4-aminophenyl-4H-imidazole-1-yl methanone at 2.3 A resolution | | Descriptor: | (4-aminophenyl)-imidazol-1-yl-methanone, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dube, D, Singh, R.P, Sinha, M, Singh, A.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-04-18 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of goat Lactoperoxidase complexed with a tightly bound inhibitor, 4-aminophenyl-4H-imidazole-1-yl methanone at 2.3 A resolution
TO BE PUBLISHED
|
|
3RL9
 
 | | Crystal Structure of the complex of type I RIP with the hydrolyzed product of dATP, adenine at 1.9 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, GLYCEROL, ... | | Authors: | Kushwaha, G.S, Pandey, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-04-19 | | Release date: | 2011-05-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a type-1 ribosome inactivating protein from Momordica balsamina in the bound and unbound states
Biochim.Biophys.Acta, 1824, 2012
|
|
