6L63
 
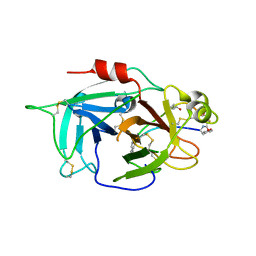 | | Human Coagulation Factor XIIa (FXIIa) bound with the macrocyclic peptide F3 containing two (1S,2S)-2-ACHC residues | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL GROUP, ... | | Authors: | Sengoku, T, Katoh, T, Hirata, K, Suga, H, Ogata, K. | | Deposit date: | 2019-10-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Ribosomal synthesis and de novo discovery of bioactive foldamer peptides containing cyclic beta-amino acids.
Nat.Chem., 12, 2020
|
|
5WXI
 
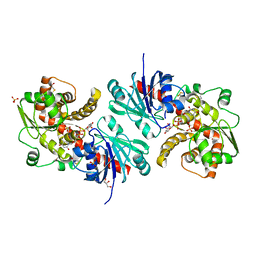 | | EarP bound with dTDP-rhamnose (soaked) | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, BETA-MERCAPTOETHANOL, EarP, ... | | Authors: | Sengoku, T, Yokoyama, S, Yanagisawa, T. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of protein arginine rhamnosylation by glycosyltransferase EarP
Nat. Chem. Biol., 14, 2018
|
|
5WXJ
 
 | | Apo EarP | | Descriptor: | BETA-MERCAPTOETHANOL, EarP, GLYCEROL, ... | | Authors: | Sengoku, T, Yokoyama, S, Yanagisawa, T. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-28 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of protein arginine rhamnosylation by glycosyltransferase EarP
Nat. Chem. Biol., 14, 2018
|
|
7X5G
 
 | | Nrf2 (A510Y)-MafG heterodimer bound with CsMBE2 | | Descriptor: | DNA (5'-D(*CP*AP*CP*AP*GP*TP*GP*AP*CP*TP*CP*AP*GP*CP*AP*G)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*TP*GP*AP*GP*TP*CP*AP*CP*TP*GP*T)-3'), Nuclear factor erythroid 2-related factor 2, ... | | Authors: | Sengoku, T, Shiina, M, Suzuki, K, Hamada, K, Sato, K, Uchiyama, A, Okada, C, Baba, S, Ohta, T, Motohashi, H, Yamamoto, M, Ogata, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Nucleic Acids Res., 50, 2022
|
|
7X5E
 
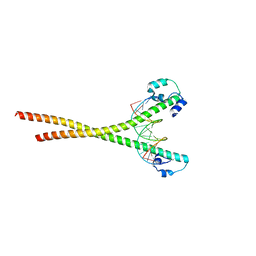 | | Nrf2-MafG heterodimer bound with CsMBE1 | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*AP*TP*GP*AP*GP*TP*CP*AP*GP*CP*AP*A)-3'), DNA (5'-D(*GP*TP*TP*GP*CP*TP*GP*AP*CP*TP*CP*AP*TP*CP*AP*T)-3'), HEXAETHYLENE GLYCOL, ... | | Authors: | Sengoku, T, Shiina, M, Suzuki, K, Hamada, K, Sato, K, Uchiyama, A, Okada, C, Baba, S, Ohta, T, Motohashi, H, Yamamoto, M, Ogata, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Nucleic Acids Res., 50, 2022
|
|
7X5F
 
 | | Nrf2-MafG heterodimer bound with CsMBE2 | | Descriptor: | Nuclear factor erythroid 2-related factor 2, Synthetic DNA, Transcription factor MafG | | Authors: | Sengoku, T, Shiina, M, Suzuki, K, Hamada, K, Sato, K, Uchiyama, A, Okada, C, Baba, S, Ohta, T, Motohashi, H, Yamamoto, M, Ogata, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Nucleic Acids Res., 50, 2022
|
|
5XVR
 
 | |
5WXK
 
 | | EarP bound with domain I of EF-P | | Descriptor: | BETA-MERCAPTOETHANOL, EarP, Elongation factor P, ... | | Authors: | Sengoku, T, Yokoyama, S, Yanagisawa, T. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis of protein arginine rhamnosylation by glycosyltransferase EarP
Nat. Chem. Biol., 14, 2018
|
|
7E8D
 
 | | NSD2 E1099K mutant bound to nucleosome | | Descriptor: | DNA (185-MER), Histone H2A type 1, Histone H2B type 1-J, ... | | Authors: | Sengoku, T, Sato, K, Nishizawa, T, Nureki, O, Ogata, K. | | Deposit date: | 2021-03-01 | | Release date: | 2021-11-10 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of the regulation of the normal and oncogenic methylation of nucleosomal histone H3 Lys36 by NSD2.
Nat Commun, 12, 2021
|
|
7FBP
 
 | | FXIIa-cMCoFx1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XIIa light chain, cMCoFx1 | | Authors: | Sengoku, T, Liu, W, de Veer, S.J, Huang, Y.H, Okada, C, Zdenek, C.N, Fry, B.G, Swedberg, J.E, Passioura, T, Craik, D.J, Suga, H, Ogata, K. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | An Ultrapotent and Selective Cyclic Peptide Inhibitor of Human beta-Factor XIIa in a Cyclotide Scaffold.
J.Am.Chem.Soc., 143, 2021
|
|
2DB3
 
 | | Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3', ATP-dependent RNA helicase vasa, MAGNESIUM ION, ... | | Authors: | Sengoku, T, Nureki, O, Nakamura, A, Kobayashi, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-14 | | Release date: | 2006-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa.
Cell(Cambridge,Mass.), 125, 2006
|
|
2DW4
 
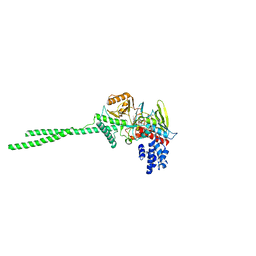 | |
2EJR
 
 | | LSD1-tranylcypromine complex | | Descriptor: | Lysine-specific histone demethylase 1, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL (2R,3S,4S)-5-[7,8-DIMETHYL-2,4-DIOXO-5-(3-PHENYLPROPANOYL)-1,3,4,5-TETRAHYDROBENZO[G]PTERIDIN-10(2H)-YL]-2,3,4-TRIHYDROXYPENTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Sengoku, T, Mimasu, S, Umehara, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-20 | | Release date: | 2008-01-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of histone demethylase LSD1 and tranylcypromine at 2.25A
Biochem.Biophys.Res.Commun., 366, 2008
|
|
3AVS
 
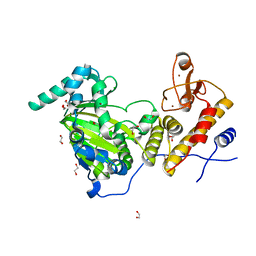 | | Catalytic fragment of UTX/KDM6A bound with N-oxyalylglycine, and Ni(II) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 6A, ... | | Authors: | Sengoku, T, Yokoyama, S. | | Deposit date: | 2011-03-07 | | Release date: | 2011-10-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for histone H3 Lys 27 demethylation by UTX/KDM6A
Genes Dev., 25, 2011
|
|
3AVR
 
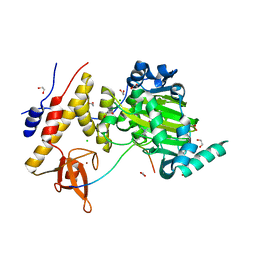 | | Catalytic fragment of UTX/KDM6A bound with histone H3K27me3 peptide, N-oxyalylglycine, and Ni(II) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Histone H3, ... | | Authors: | Sengoku, T, Yokoyama, S. | | Deposit date: | 2011-03-07 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural basis for histone H3 Lys 27 demethylation by UTX/KDM6A
Genes Dev., 25, 2011
|
|
8YE0
 
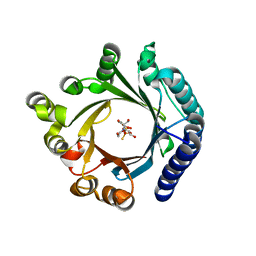 | | Crystal structure of KgpF prenyltransferase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LynF/TruF/PatF family peptide O-prenyltransferase, MAGNESIUM ION, ... | | Authors: | Hamada, K, Inoue, S, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2024-02-21 | | Release date: | 2024-06-19 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | De Novo Discovery of Pseudo-Natural Prenylated Macrocyclic Peptide Ligands.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8WM0
 
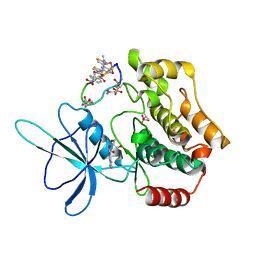 | | Crystal structure of TNIK-thiopeptide wTP3 complex | | Descriptor: | ADENOSINE, THIOPEPTIDE wTP3, TRAF2 and NCK-interacting protein kinase | | Authors: | Hamada, K, Kobayashi, S, Vinogradov, A.A, Zhang, Y, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2023-10-01 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Compact Reprogrammed Genetic Code for De Novo Discovery of Proteolytically Stable Thiopeptides.
J.Am.Chem.Soc., 2024
|
|
7VMY
 
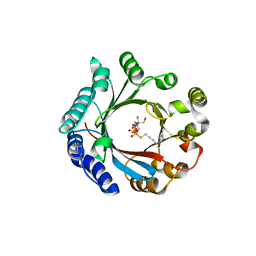 | | Crystal structure of LimF prenyltransferase bound with GSPP | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GERANYL S-THIOLODIPHOSPHATE, LynF/TruF/PatF family peptide O-prenyltransferase, ... | | Authors: | Hamada, K, Kobayashi, S, Okada, C, Zhang, Y, Inoue, S, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | LimF is a versatile prenyltransferase for histidine-C-geranylation on diverse non-natural substrates
Nat Catal, 2022
|
|
7VMW
 
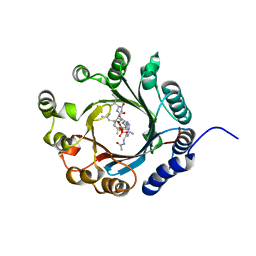 | | Crystal structure of LimF prenyltransferase bound with a peptide substrate and GSPP | | Descriptor: | GERANYL S-THIOLODIPHOSPHATE, LynF/TruF/PatF family peptide O-prenyltransferase, MAGNESIUM ION, ... | | Authors: | Hamada, K, Kobayashi, S, Okada, C, Zhang, Y, Inoue, S, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | LimF is a versatile prenyltransferase for histidine-C-geranylation on diverse non-natural substrates
Nat Catal, 2022
|
|
7XZR
 
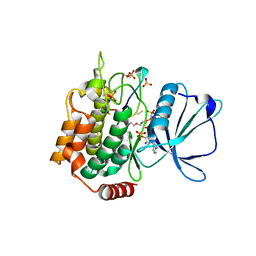 | | Crystal structure of TNIK-AMPPNP-thiopeptide TP15 complex | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION, ... | | Authors: | Hamada, K, Vinogradov, A.A, Zhang, Y, Chang, J.S, Nishimura, H, Goto, Y, Onaka, H, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | De Novo Discovery of Thiopeptide Pseudo-natural Products Acting as Potent and Selective TNIK Kinase Inhibitors.
J.Am.Chem.Soc., 144, 2022
|
|
7XZQ
 
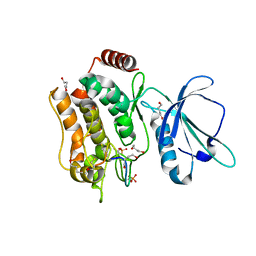 | | Crystal structure of TNIK-thiopeptide TP1 complex | | Descriptor: | 1,4-BUTANEDIOL, TRAF2 and NCK-interacting protein kinase, thiopeptide TP1 | | Authors: | Hamada, K, Vinogradov, A.A, Zhang, Y, Chang, J.S, Nishimura, H, Goto, Y, Onaka, H, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | De Novo Discovery of Thiopeptide Pseudo-natural Products Acting as Potent and Selective TNIK Kinase Inhibitors.
J.Am.Chem.Soc., 144, 2022
|
|
1ULH
 
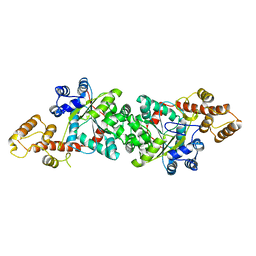 | | A short peptide insertion crucial for angiostatic activity of human tryptophanyl-tRNA synthetase | | Descriptor: | Tryptophanyl-tRNA synthetase | | Authors: | Kise, Y, Sengoku, T, Ishii, R, Yokoyama, S, Park, S.G, Lee, S.W, Kim, S, Nureki, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-12 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A short peptide insertion crucial for angiostatic activity of human tryptophanyl-tRNA synthetase
Nat.Struct.Mol.Biol., 11, 2004
|
|
1WWR
 
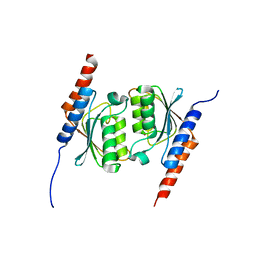 | | Crystal structure of tRNA adenosine deaminase TadA from Aquifex aeolicus | | Descriptor: | ZINC ION, tRNA adenosine deaminase TadA | | Authors: | Kuratani, M, Ishii, R, Bessho, Y, Fukunaga, R, Sengoku, T, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-12 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of tRNA Adenosine Deaminase (TadA) from Aquifex aeolicus
J.Biol.Chem., 280, 2005
|
|
8GUJ
 
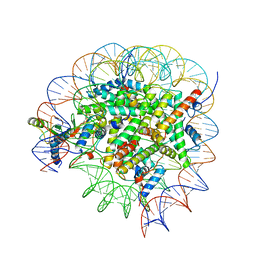 | | Bre1-nucleosome complex (Model II) | | Descriptor: | DNA (147-mer), E3 ubiquitin-protein ligase BRE1A, E3 ubiquitin-protein ligase BRE1B, ... | | Authors: | Onishi, S, Sato, K, Hamada, K, Nishizawa, T, Nureki, O, Ogata, K, Sengoku, T. | | Deposit date: | 2022-09-12 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the human Bre1 complex bound to the nucleosome.
Nat Commun, 15, 2024
|
|
8GUK
 
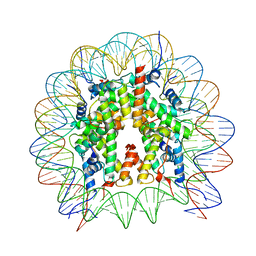 | | Human nucleosome core particle (free form) | | Descriptor: | DNA (147-mer), Histone H2A type 1, Histone H2B type 1-J, ... | | Authors: | Onishi, S, Sato, K, Nishizawa, T, Nureki, O, Ogata, K, Sengoku, T. | | Deposit date: | 2022-09-12 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Structure of the human Bre1 complex bound to the nucleosome.
Nat Commun, 15, 2024
|
|
