4B33
 
 | | Humanised monomeric RadA in complex with napht-2-ol | | Descriptor: | 1-NAPHTHOL, DNA REPAIR AND RECOMBINATION PROTEIN RADA, PHOSPHATE ION | | Authors: | Scott, D.E, Ehebauer, M.T, Pukala, T, Marsh, M, Blundell, T.L, Venkitaraman, A.R, Abell, C, Hyvonen, M. | | Deposit date: | 2012-07-20 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Using a Fragment-Based Approach to Target Protein-Protein Interactions.
Chembiochem, 14, 2013
|
|
3TDI
 
 | | yeast Cul1WHB-Dcn1P acetylated Ubc12N complex | | Descriptor: | Defective in cullin neddylation protein 1, NEDD8-conjugating enzyme UBC12 | | Authors: | Scott, D.C, Monda, J.K, Bennett, E.J, Harper, J.W, Schulman, B.A. | | Deposit date: | 2011-08-11 | | Release date: | 2011-10-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex.
Science, 334, 2011
|
|
3TDZ
 
 | | N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex: Structure of a human Cul1WHB-Dcn1P-stapled acetylated Ubc12N complex | | Descriptor: | Cullin-1, DCN1-like protein 1, STAPLED PEPTIDE | | Authors: | Scott, D.C, Monda, J.K, Bennett, E.J, Harper, J.W, Schulman, B.A. | | Deposit date: | 2011-08-11 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex.
Science, 334, 2011
|
|
3TDU
 
 | | N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex: Structure of a human Cul1WHB-Dcn1P-acetylated Ubc12N complex | | Descriptor: | Cullin-1, DCN1-like protein 1, NEDD8-conjugating enzyme Ubc12 | | Authors: | Scott, D.C, Monda, J.K, Bennett, E.J, Harper, J.W, Schulman, B.A. | | Deposit date: | 2011-08-11 | | Release date: | 2011-10-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex.
Science, 334, 2011
|
|
4P5O
 
 | |
5J4K
 
 | | Structure of humanised RadA-mutant humRadA22F in complex with 1-Indane-6-carboxylic acid | | Descriptor: | 2,3-dihydro-1H-indene-2-carboxylic acid, CALCIUM ION, DNA repair and recombination protein RadA, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-01 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.346 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5JEC
 
 | | Apo-structure of humanised RadA-mutant humRadA33F | | Descriptor: | CHLORIDE ION, DNA repair and recombination protein RadA, SULFATE ION | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-18 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5JED
 
 | | Apo-structure of humanised RadA-mutant humRadA28 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-18 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5J4H
 
 | | Structure of humanised RadA-mutant humRadA22F in complex with indole-6-carboxylic acid | | Descriptor: | 1H-indole-6-carboxylic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-01 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
4UQO
 
 | | RADA C-TERMINAL ATPASE DOMAIN FROM PYROCOCCUS FURIOSUS BOUND TO ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA REPAIR AND RECOMBINATION PROTEIN RADA, MAGNESIUM ION, ... | | Authors: | Marsh, M.E, Ehebauer, M.T, Scott, D, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2014-06-24 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | ATP Half-Sites in Rada and Rad51 Recombinases Bind Nucleotides
FEBS Open Bio, 6, 2016
|
|
6TW3
 
 | | HumRadA2 in complex with Naphthyl-HPA fragment-peptide chimera | | Descriptor: | (2~{S})-1-[(2~{S})-2-[(3-azanylnaphthalen-2-yl)carbonylamino]-3-(1~{H}-imidazol-4-yl)propanoyl]-~{N}-[(2~{S})-1-azanyl-1-oxidanylidene-propan-2-yl]pyrrolidine-2-carboxamide, DNA repair and recombination protein RadA, PHOSPHATE ION | | Authors: | Marsh, M.E, Fischer, G, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.352 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
6TW4
 
 | | HumRadA22F in complex with compound 6 | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA, ~{N}-[2-[(2~{S})-2-[[(1~{S})-1-(4-methoxyphenyl)ethyl]carbamoyl]pyrrolidin-1-yl]-2-oxidanylidene-ethyl]quinoline-2-carboxamide | | Authors: | Marsh, M.E, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
6TW9
 
 | | HumRadA22F in complex with CAM833 | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA, GLYCEROL, ... | | Authors: | Fischer, G, Marsh, M.E, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | Deposit date: | 2020-01-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
7KWA
 
 | | Structure of DCN1 bound to N-((4S,5S)-3-(aminomethyl)-7-ethyl-4-(4-fluorophenyl)-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)-3-(trifluoromethyl)benzamide | | Descriptor: | Endolysin,DCN1-like protein 1, N-[(4S,5S)-3-(aminomethyl)-7-ethyl-4-(4-fluorophenyl)-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]-3-(trifluoromethyl)benzamide | | Authors: | Kim, H.S, Hammill, J.T, Schulman, B.A, Guy, R.K, Scott, D.C. | | Deposit date: | 2020-11-30 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.572 Å) | | Cite: | Improvement of Oral Bioavailability of Pyrazolo-Pyridone Inhibitors of the Interaction of DCN1/2 and UBE2M.
J.Med.Chem., 64, 2021
|
|
6BG5
 
 | | Structure of 1-(benzo[d][1,3]dioxol-5-ylmethyl)-1-(1-propylpiperidin-4-yl)-3-(3-(trifluoromethyl)phenyl)urea bound to DCN1 | | Descriptor: | Endolysin, DCN1-like protein 1 chimera, N-[(2H-1,3-benzodioxol-5-yl)methyl]-N-(1-propylpiperidin-4-yl)-N'-[3-(trifluoromethyl)phenyl]urea | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-10-27 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Piperidinyl Ureas Chemically Control Defective in Cullin Neddylation 1 (DCN1)-Mediated Cullin Neddylation.
J. Med. Chem., 61, 2018
|
|
6P5W
 
 | | Structure of DCN1 bound to 3-methyl-N-((4S,5S)-3-methyl-6-oxo-1-phenyl-4-(p-tolyl)-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)benzamide | | Descriptor: | 3-methyl-N-[(4S,5S)-3-methyl-4-(4-methylphenyl)-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]benzamide, Lysozyme,DCN1-like protein 1 chimera | | Authors: | Guy, R.K, Kim, H.S, Hammill, J.T, Scott, D.C, Schulman, B.A. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of Novel Pyrazolo-pyridone DCN1 Inhibitors Controlling Cullin Neddylation.
J.Med.Chem., 62, 2019
|
|
6V4T
 
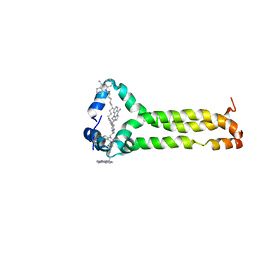 | | MPER-TMD of HIV-1 Env bound with the entry inhibitor S2C3 | | Descriptor: | 4,4'-(decane-1,10-diyl)bis(9-amino-2,3-dihydro-1H-cyclopenta[b]quinolin-4-ium), Envelope glycoprotein gp160 | | Authors: | Xiao, T, Frey, G, Fu, Q, Lavine, C.L, Scott, D.A, Seaman, M.S, Chou, J.J, Chen, B. | | Deposit date: | 2019-11-30 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | HIV-1 fusion inhibitors targeting the membrane-proximal external region of Env spikes.
Nat.Chem.Biol., 16, 2020
|
|
6P5V
 
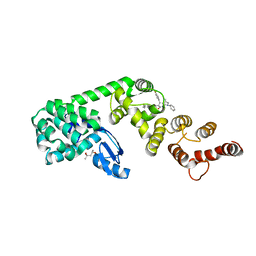 | | Structure of DCN1 bound to N-((4S,5S)-7-ethyl-4-(4-fluorophenyl)-3-methyl-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)-3-methylbenzamide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lysozyme,DCN1-like protein 1 fusion, N-[(4S,5S)-1-[(1S)-cyclohex-3-en-1-yl]-7-ethyl-4-(4-fluorophenyl)-3-methyl-6-oxo-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]-3-methylbenzamide | | Authors: | Guy, R.K, Kim, H.S, Hammill, J.T, Scott, D.C, Schulman, B.A. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Discovery of Novel Pyrazolo-pyridone DCN1 Inhibitors Controlling Cullin Neddylation.
J.Med.Chem., 62, 2019
|
|
2N5G
 
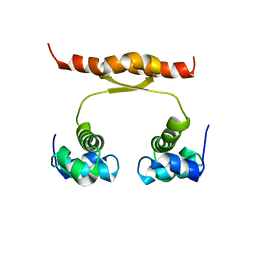 | | NMR structure of KorA, a plasmid-encoded, global transcription regulator KorA | | Descriptor: | TrfB transcriptional repressor protein | | Authors: | Rajasekar, K.V, Lovering, A.L, Dancea, F.V, Scott, D.J, Harris, S, Bingle, L.E, Roessle, M, Thomas, C.M, Hyde, E.I, White, S.A. | | Deposit date: | 2015-07-17 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Flexibility of KorA, a plasmid-encoded, global transcription regulator, in the presence and the absence of its operator.
Nucleic Acids Res., 44, 2016
|
|
5JEE
 
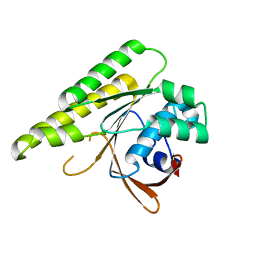 | | Apo-structure of humanised RadA-mutant humRadA26F | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-18 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5L8V
 
 | | Apo-structure of humanised RadA-mutant humRadA4 | | Descriptor: | DNA repair and recombination protein RadA, PHOSPHATE ION | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-08 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5LB4
 
 | | Apo-structure of humanised RadA-mutant humRadA14 | | Descriptor: | DNA repair and recombination protein RadA | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-15 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5LBI
 
 | | Apo-structure of humanised RadA-mutant humRadA3 | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-16 | | Release date: | 2016-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5LB2
 
 | | Apo-structure of humanised RadA-mutant humRadA2 | | Descriptor: | DNA repair and recombination protein RadA | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-15 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
1UPF
 
 | | STRUCTURE OF THE URACIL PHOSPHORIBOSYLTRANSFERASE, MUTANT C128V BOUND TO THE DRUG 5-FLUOROURACIL | | Descriptor: | 5-FLUOROURACIL, SULFATE ION, URACIL PHOSPHORIBOSYLTRANSFERASE | | Authors: | Schumacher, M.A, Carter, D, Scott, D, Roos, D, Ullman, B, Brennan, R.G. | | Deposit date: | 1998-06-17 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding.
EMBO J., 17, 1998
|
|
